《1 Introduction》
1 Introduction
Microorganisms are ubiquitous and closely related to production, human survival, and life. They are widely used for the production of food, such as cheese, wine, yogurt, bread, and steamed bread. In addition, they can also be used for the purification of water sources and the adsorption of heavy metals. The intestinal microorganisms in the human body help to degrade and absorb food. The existence of microorganisms connects all aspects of natural order and human life. Microorganisms are closely related to human health, and most of these organisms are harmless and can coexist peacefully with their hosts. However, some microorganisms, namely pathogenic microorganisms, can cause harm to humans, animals, or plants. Pathogenic microorganisms can cause host infection, allergies, tumors, dementia, and even death, and they are also one of the main factors endangering food safety. In recent years, severe acute respiratory syndrome, highly pathogenic avian influenza, West Nile virus infection, new coronavirus pneumonia, and other diseases have been identified to have the characteristics of strong infectivity and have caused great harm to human body. Therefore, to ensure human health, food safety, and food security, it is necessary to carry out rapid, accurate, and sensitive detection and accurate classification of microorganisms in the fields of medicine, environmental science, agriculture, and food. The traditional microbial tests, including common pathogenic bacteria test processes, such as colony morphology, physiological and biochemical indexes, and serotype detection with mature technology and simple equipment, are still the mainstream detection methods adopted by food health regulatory agencies. However, the experimental operation steps are cumbersome and time-consuming, the methods have low sensitivity, and the test results lack sufficient persuasion.
With the development of medical microbiology research technology and interdisciplinary methods, new microbial detection technologies have emerged, overcoming one or more limitations of traditional methods of detecting and identifying microorganisms. The current microbial detection technology has advanced to the molecular and gene levels with development of different technologies, such as nucleic acid hybridization technology, nucleic acid amplification technology, deoxyribonucleic acid (DNA) fingerprint profiling technology, gene chip technology, and high-throughput sequencing technology. With the development of technology and automation equipment, the rapidity, sensitivity, specificity, and applicability of microbial detection have significantly improved. However, it should be noted that each microbial detection method has its own advantages and limitations in one or more aspects, with regard to the duration of analysis, sensitivity, stability, and experimental environment, which limit the application. In view of this, this paper highlights the developmental progress and application trend of molecular identification methods of microbial resources, analyzes the existing problems, and puts forward suggestions for the development of molecular identification methods of microbial resources in China to provide basic reference for relevant research and development layout as well as research management policy.
《2 Outline of the molecular identification methods of microbial resources》
2 Outline of the molecular identification methods of microbial resources
The community characteristics of microorganisms pose a challenge to microbial detection technology, not only for microbial detection, but also for the detection of variations and classification of microorganisms. Molecular identification of microbial resources involves classifying and locating the biological evolution diversity of natural microbial resources by using the conserved genome sequences and the subtle changes and laws preserved in the long-term evolution of microorganisms [1]. Molecular identification has become necessary classification information in microbial resource identification, and it is also an important method for rapid identification of microbial resources. Common molecular identification techniques include the following:
《2.1 DNA (G+C) content (mol%) identification》
2.1 DNA (G+C) content (mol%) identification
The specificity of genetic material DNA is determined by the arrangement and content of adenine (A), guanine (G), thymine (T), and cytosine (C). The (G+C) content of DNA differs in different organisms and is usually expressed as the mol% content of (G+C) in DNA [2]. It is species-specific and not affected by bacterial age and external factors, and hence, it has become an important indicator for bacterial classification and strain identification. The (G+C) content of different microbial DNA varies widely, but the (G+C) content of the same microorganism fluctuates only between 3% and 5%. Therefore, DNA (G+C) content (mol%) identification method can be used for rapid detection of microbial population. Microorganisms closely related or highly similar in phenotype have similar (G+C) content, but not exactly the same content; the difference in (G+C) content between the same genus bacteria is ≤ 12 mol%, and the difference between strains of the same species is ˂ 5 mol%. Therefore, the genetic relationship between bacteria can be judged by data difference. It should be noted that this identification method is mainly used to exclude uncertain classification units, rather than to establish a new classification unit, that is, for species with close genetic relationship, the content of (G+C) is similar, but species with similar (G+C) content may not necessarily have a close genetic relationship.
《2.2 Nucleic acid hybridization technique》
2.2 Nucleic acid hybridization technique
Nucleic acid hybridization is the earliest developed molecular biological technology used for microbial detection, which involves the hybridization of nucleic acid probes of known sequences and markers with target sequences, and the detection of specific binding probes with specific methods [3]. Each microorganism has a few nucleic acid fragments that are specific to it. After separation and labeling, probes are prepared, and the DNA sequence fragments are analyzed using qualitative and quantitative detection probe for microbial identification. This technique has the advantages of simple operation, good specificity, high sensitivity, and fast speed. Its main types are imprinting hybridization, spot hybridization, and in situ hybridization. Among these techniques, fluorescence in situ hybridization (FISH), developed on the basis of in situ hybridization, has been widely used with fluorescently labeled probes and fluorescence microscopy. However, this method is susceptible to hybridization temperature, reaction conditions, fluorescent dyes, and probe selection. It should be pointed out that nucleic acid hybridization technology requires the use of radioactive isotopes, which are costly and will cause certain harm to human body, limiting the application and development of the technology to a certain extent. However, in combination with biosensor technology, the benefits are significantly improved.
《2.3 Nucleic acid amplification technique》
2.3 Nucleic acid amplification technique
2.3.1 Polymerase chain reaction (PCR) technique
PCR technology is one of the most commonly used molecular methods and also one of the commonly used detection techniques in the current national and industry standards for microbial detection. PCR technology amplifies a specific target DNA sequence through a two-step or three-step temperature cycle, which includes template denaturation, primer annealing, and product extension.
According to the number of the detection targets, PCR-based microbial detection technology can be divided into single-PCR technology and multiple-PCR technology [4]. (1) Single PCR technology refers to the detection method that involves the amplification of only one pair of primers in one PCR reaction, which is suitable for the detection of highly conserved microbial species or genera. By detecting the species-specific and intraspecific conserved DNA sequences of the target microorganisms, the existence of the target microorganisms is confirmed by the detection results; for example, the detection of Pantoea in onion [5] and pathogenic bacteria in eggplant [6]. Single PCR can only detect one target at a time, as it has the limitations of low detection efficiency, high false negative, and low sensitivity caused by single primer amplification failure. (2) Multiplex PCR technology refers to a PCR reaction that involves the amplification of more than one pair of primers, which can detect multiple targets. Multiplex PCR is a method that can detect multiple targets with PCR amplification. This technology can detect multiple target microorganisms or multiple target sequences of a target microorganism at a time, thus exhibiting significant detection efficiency. The detection of multiple target sequences of a target microorganism significantly improves the accuracy of detection. Multiplex PCR technology has been used for the detection of multiple serotypes of Salmonella [7], fungal microorganisms [4], ocular bacterial pathogens [8], 18 pathogens in cerebrospinal fluid of children with viral meningitis [9], 15 intestinal pathogens in fecal samples [10], and 11 food-borne pathogens [11].
PCR-based detection technology also includes droplet PCR [12], which integrates PCR system into droplets, and solid-phase PCR [7], which solidifies PCR primers on fixed support. However, due to the complex operation and high cost, these PCR techniques are not often used for practical applications.
In practical application, fluorescent PCR technology is more commonly used, including in China, for the detection of pathogenic microorganisms [13]. The discovery of fluorescent substances simplifies the detection steps of PCR products. Fluorescent substances are added to the single or multiple PCR reaction systems, and the formation of PCR products is monitored in real time through the accumulation of fluorescent signals. Therefore, the addition of fluorescent substances facilitates the real-time quantitative detection of target microorganisms by PCR technology. Fluorescence PCR technology is usually combined with multiplex PCR for real-time quantitative detection of multiple targets. Common fluorescent substances are fluorescent dyes (such as SYBR Green nucleic acid dye) and fluorescent probes (such as Taqman probe). For example, a variety of bacterial pathogens in the eyes [8] and 15 intestinal pathogens in feces samples [10] were detected based on the SYBR Green method. The Taqman probe method has been used to detect Candida spp., including the multi-resistant Candida spp. [7,14], and a novel coronavirus [15,16]. At present, the number of primers required for one-time multiplex PCR detection based on fluorescence PCR instrument is low. When used for simultaneous detection of multiple microorganisms, the technique can detect only one target sequence per microorganism; hence, there is a chance of false negative detection which means that the microorganism is not detected because of a sequence detection failure.
2.3.2 Isothermal amplification technique
Isothermal amplification technology is the amplification of nucleic acid under isothermal conditions and is widely used in the molecular diagnosis of grassroots and instant microorganisms [17,18]. Based on the different active enzymes added, the common isothermal amplification technologies include loop-mediated isothermal nucleic acid amplification (LAMP), rolling-loop amplification, single-primer isothermal amplification, and helicase-dependent isothermal amplification. Isothermal amplification technology has the following characteristics: (1) simple operation compared to conventional PCR technology; no template thermal degeneration, electrophoresis, and UV observation required, as product detection results can be judged by naked eye or turbidity meter by detecting precipitation turbidity; (2) no requirement of temperature cycle and high speed; (3) extremely high specificity, as no nucleic acid amplification can be performed in any of the six target regions that do not match the primers; (4) high sensitivity, as detection can be achieved with low number of copies of viral template, indicating its advantage over the PCR technology. Isothermal amplification technology has been widely used for the detection of different pathogens [19], including apple rot pathogen [20], potato ring rot [21], sugarcane ratoon dwarf pathogen [22] and other plant pathogens, Streptococcus [23] and Lactococcus in fish [24], and other animal and human respiratory pathogens [25].
《2.4 DNA fingerprinting technique》
2.4 DNA fingerprinting technique
DNA fingerprint technology uses DNA fingerprint to classify DNA sequence and identify microorganisms at the species level. DNA-based genotyping method is simple and easy to carry out, with high repeatability and resolution [26]. DNA fingerprinting technology (1) is multi-locus, indicating that it can provide comprehensive characteristics of the genome; (2) has high variability, indicating that different species have different DNA profiles; (3) is simple and has stable heredity, that is, DNA maps can be accurately passed from the previous generation to the next generation. At present, this technology has become a conventional method for identification and classification research, which is not only used for classification and the determination of diversity and systematics of microorganisms at the different levels, but also for elucidating the evolutionary relationship and ecology of microorganisms.
The basic method of DNA fingerprinting is to digest extracted genomic DNA, separate it by agarose gel electrophoresis, perform southern blotting, and finally obtain DNA fingerprints. After a series of analysis of the fingerprint, the corresponding information can be obtained to determine the genetic relationship of the strains. DNA fingerprinting techniques include denaturing gradient gel electrophoresis, temperature gradient gel electrophoresis, pulsed field gel electrophoresis (PFGE), random amplified polymorphic DNA, restriction fragment length polymorphism, repetitive sequence PCR, variable number tandem repeat analysis (MLVA), and multi-locus sequence analysis (MLST). DNA fingerprinting technology based on PCR technology is a commonly used method because of the benefit of combining the two technologies, and the method does not need a gene library. The application characteristics of methods using this technology are different and thus they can be applied for identifying different microbes according to the situation. Therefore, two methods are usually used together in specific experiments. However, there are inevitably some shortcomings.
《2.5 Gene chip technology》
2.5 Gene chip technology
Gene chip technology is a biochip technology that uses hybridization technology to achieve genetic detection. The technique is a high-tech technology that regularly arranges and anchors tens of thousands of sequence-specific DNA fragments to proponents, constituting a two-dimensional DNA probe array. Because the chip is similar to the electronic chip of the computer, it is called the gene chip technology [27]. Due to the strong specificity and high throughput of chip technology, it is suitable for a variety of microorganisms in one-time detection samples. After the test results are obtained, they can be automatically analyzed based on biological knowledge. Gene chip technology plays an important role in microbial pathogen detection, species identification, functional gene detection, genotyping, mutation detection, and genome monitoring [28].
Gene chip technology can achieve high-throughput detection with high detection efficiency. However, this technology has the problems of large background noise and low sensitivity associated with coloration or fluorescence signals. Although it can detect the presence or absence of target microorganisms, it cannot detect sequence variation and classify microorganisms. In addition, most of the existing chips are dot-array hybrid chips, and although some chips are commercialized, they still cannot meet the needs of practical applications. Other challenges associated with the chip are limited function, the need for customization, high cost, and poor flexibility.
《2.6 High-throughput sequencing》
2.6 High-throughput sequencing
The first generation sequencing technology was developed based on the Sanger method, and the next generation sequencing (NGS) technology was later developed. The NGS method has been able to perform genome-wide sequencing rapidly and efficiently. The third-generation sequencing technology developed in 2011 has a sequence reading length of up to 3000 bp. Compared with the previous two generations of sequencing technology, its biggest advantage is that it does not need PCR amplification, and hence does not involve errors caused by PCR [29]. Thanks to the continuous development of sequencing technology, millions of PCR amplicons can be sequenced simultaneously with improved detection efficiency.
2.6.1 Whole-genome sequencing (WGS), metagenomic sequencing (mNGS), and targeted sequencing techniques
WGS can accurately distinguish the genomic differences between strains and has high reproducibility. It is a powerful tool for the monitoring and investigation of bacterial isolates. WGS is not only used for expected monocytic growth monitoring, but also for the reporting of other public health-related pathogens. WGS can detect the whole genome sequence of microorganisms, and the information obtained is very comprehensive. However, this method depends on the isolation and culture of microorganisms, which is time-consuming and laborious. Because of the population characteristics of microorganisms, deep sequencing is needed for accurate detection, which makes WGS an expensive technology.
Metagenome refers to the total microbial genome in the environment. The mNGS technology does not perform single sequencing of specific microbial populations (fungi, bacteria, or viruses), but considers the entire microbial genome [30,31]. Therefore, there will be a large number of background data in the data generated by sequencing, and the quality control [32] and scientific analysis [33] of these data have become the key and difficult points for the effective use of mNGS data. In addition, another limitation of mNGS is that the target microorganism is submerged in a large amount of background data, especially when the target microorganism content is low, and it needs to be detected by deepening the sequencing depth, resulting in higher costs.
Target sequencing technology can achieve ultra-deep sequencing at low cost for specific microbial populations [34]. In terms of accuracy and sensitivity, it is significantly improved compared to mNGS technology, and hence, it can complement the advantages of mNGS technology and detect microorganisms comprehensively and accurately. Common targeted sequencing technologies include 16SrRNA, 18SrRNA, and ribosomal intergenic spacer analysis. 16SrRNA is the most widely used marker gene in species identification and taxonomy classification of bacteria and archaea [35]. 18SrRNA is a common phylogenetic gene in fungi, which has more hypervariable domains than 16SrRNA [36]. The internal transcribed spacer (ITS) between genes has attracted widespread attention because it contains good variable and conserved regions. It has outstanding advantages in the identification of species and subspecies. It is a widely used fungal barcode marker and can be used for the successful identification of fungi. 16SrRNA-, 18SrRNA-, and ITS-targeted sequencing technology can not only detect microorganisms at low cost but also classify microorganisms at the genus or species level.
2.6.2 Polynucleotide polymorphism (MNP) maker
MNP marker is a new microbial identification and DNA marker method involving the use of multiple dispersed nucleotide polymorphism markers. At present, MNP markers have been used as the technical standard of genotyping for national plant identification in China. This method involves microbial genetic variation sequencing technology (MGV-Seq) and its supporting bioinformatics analysis tools. More importantly, a microbial identification program based on MGV-Seq has been designed, and a customized calculation and statistical algorithm has been developed for MNP genotyping. MGV-Seq method has the characteristics of high reproducibility, high accuracy, high sensitivity, and high specificity. The detection of Xoo strain proved the applicability, feasibility, and reproducibility of MGV-Seq for microbial identification. Xoo strains have been distributed to laboratories around the world and have been continuously cultured for many years since they were isolated in 1972. When MGV-Seq was used to detect Xoo strains, it was unexpectedly found that some strains labeled as Xoo do not belong to the Xoo family but the Xcc family. Moreover, the reported Xoo strain may actually be a different strain.
《3 Problems associated with the molecular identification techniques of microbial resources》
3 Problems associated with the molecular identification techniques of microbial resources
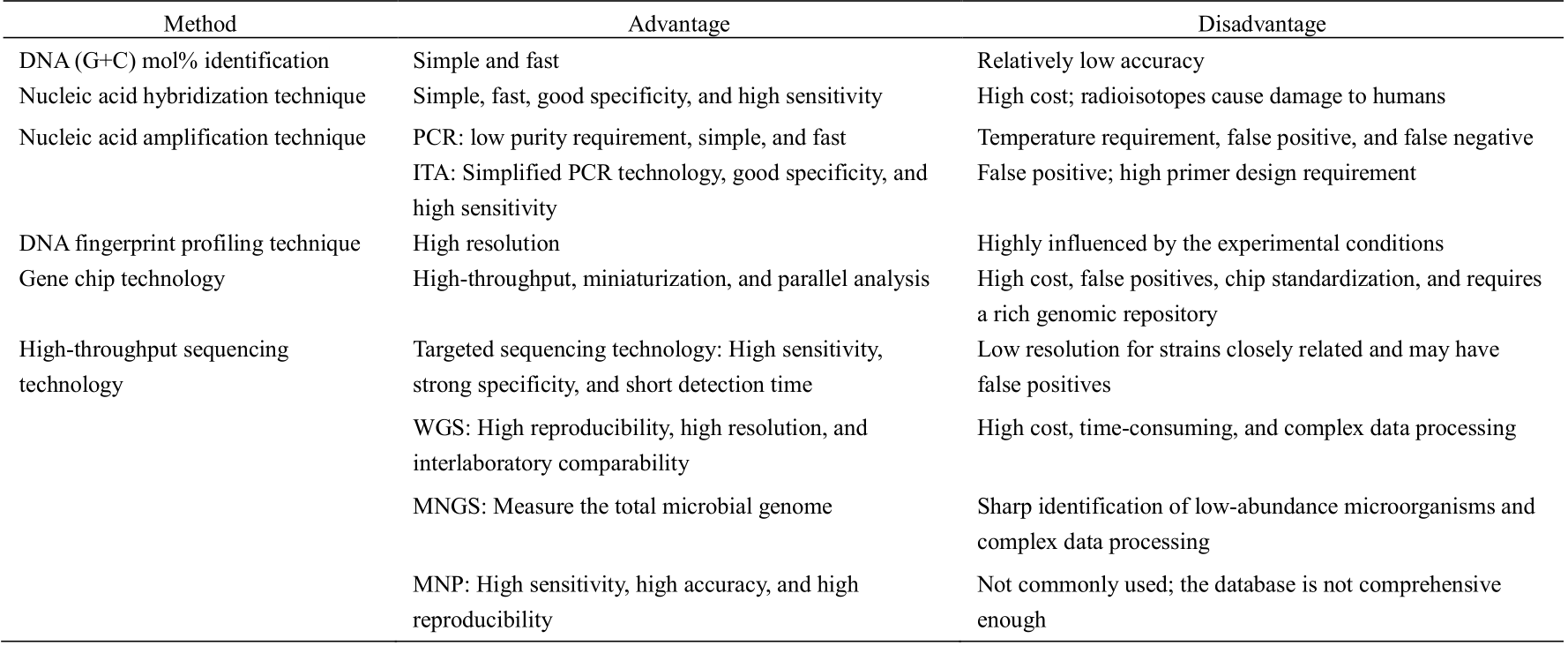
Lack of rapid and accurate detection and identification of microorganisms, especially pathogenic microorganisms, is the primary problem in food quarantine, clinical testing, and infectious disease prevention and control. The progress in molecular biology technology has advanced the development of microbial detection methods. The detection level has advanced from the histomorphology level to the molecular and the gene levels, and the detection mode has advanced from a single targeted detection mode for possible pathogens to a detection mode covering common pathogen combinations, enabling the diagnosis or exclusion of multiple pathogens in a single sample. Different molecular identification techniques for microbial resources have different applicability, advantages, and disadvantages, as shown in Table 1. To the best of our knowledge, there is currently no study on the selection of identification methods for microbial resources. To select any method, the identification cost, reproducibility, reliability, and ability to distinguish between strains need to be considered.
《Table 1》
Table 1. Advantages and disadvantages of different identification techniques
《3.1 Reliance on the laboratory environment》
3.1 Reliance on the laboratory environment
At present, the experimental methods for molecular identification of microbial resources are mostly at the laboratory stage, being only used as reference for standard detection methods. The development of identification methods that are independent of the laboratory environment, such as kits for rapid identification, is urgently needed. Moreover, microbial identification techniques depend on culture enrichment for improved detection rate, which leads to prolonged detection and experiment time. In the future, it is necessary to develop identification methods that do not require culture enrichment or that reduce enrichment time to improve detection efficiency. Furthermore, high-throughput sequencing methods are suitable for laboratory research and can obtain accurate test results, but there are some limitations, such as complex data processing and the inability to quickly obtain test results in practical applications. For a method that is too dependent on the laboratory environment, different laboratories and experimental personnel will obtain widely varied test results. Therefore, it is imperative to unify and standardize the test results to facilitate sharing and applicability of experimental results.
《3.2 Insufficient reproducibility and accuracy of experimental results》
3.2 Insufficient reproducibility and accuracy of experimental results
Microorganisms are able to rapidly reproduce and survive, making genetic variation ubiquitous in these organisms, for which they have been widely used in biological and clinical research. Genetic variation in pathogenic microorganisms can alter viral virulence, confer resistance, reduce vaccine efficacy, and significantly affect the microorganism’s interaction with the host. Genetic variation in homologous strains puts the reproducibility of experimental results at risk. To achieve the reproducibility of the experimental results, it is necessary to ensure that the same microorganism is used for the same experimental purposes in multiple experiments. Microbial identification requires the detection of genetic variation within strains and between strains. Existing microbial variant detection methods, such as PFGE, MLVA, and MLST, cannot clearly distinguish closely related strains. Generally, increased sequencing depth, improved sequencing library preparation methods, and optimized bioinformatics analysis methods, such as ultra-deep WGS, can detect rare variants, but these techniques are associated with high cost. Accuracy, as an important index for the evaluation of microbial identification technology, should be considered mainly when selecting identification methods. The existing molecular identification technology for microbial resources cannot obtain accurate results; for example, DNA (G+C) mol% identification method, which only relies on (G+C) content, cannot accurately determine microbial strain properties. PCR and gene chip technologies are associated with false negative detection, and hence, the requirements for optimizing experimental conditions or operation are relatively high. Although WGS technology can get very accurate results, the detection cost is high.
《3.3 Lack of corresponding microbial database and identification platform》
3.3 Lack of corresponding microbial database and identification platform
At present, the database construction of genomic information of microbial resources in China is in the initial stage. The relevant microbial genomics, metabolomics, transcriptomics, proteomics and other information depend on foreign databases, and the database of the U.S. National Center for Biotechnology Information occupies the dominant position. For example, when using gene chip technology for identification, specific genes or ribosomal gene information of bacteria are needed to design target genes, and hence, a perfect genomic database is particularly important. Applied microbial DNA identification platform mainly relies on 16SrRNA genes, ITS genes, and allogeneic identification techniques. 16SrRNA and ITS gene identification techniques can only distinguish the classification level of species or genera and cannot accurately distinguish harmful and beneficial microbes. Specific gene identification technology can only identify the species or races of ˂ 10 kinds of microorganisms at a time and cannot detect multiple microorganisms in food at the same time. In addition, in order to avoid aerosol pollution, this identification technology has high requirements for the laboratory environment and personnel. The identification of microbial resources based on 16SrRNA, ITS, and other technologies is highly dependent on foreign equipment, such as those from Illumina and Thermo Fisher Scientific, which have some disadvantages, such as high cost and expensive consumables, and unprotected intellectual property rights of identification technology. The bottleneck of high-end reagents and high-end instruments and equipment is serious; hence, the establishment of a microbial DNA identification platform with independent intellectual property rights is urgently required.
《4 Development proposal》
4 Development proposal
Microbial classification and identification has become an important part of microbiology research. It does not only clarify the biological status and essence of microbial resources, but it is also an important step to realize the utilization of microbial resources. At present, due to the lack of accurate microbial resource identification methods, the phenomenon of stealing microbial strains has occurred from time to time, which not only causes certain economic losses to the production enterprises, but also causes irreparable losses to the researchers who spent years or even decades on microbial strains, seriously affecting the development of microbial health and safety industries. Therefore, accurate identification of microbial resources is of great significance to the screening of microbial germplasm and protection of intellectual property rights. To promote the protection and utilization of microbial resources in China, the following recommendations are proposed.
《4.1 Continuous research on each identification technology》
4.1 Continuous research on each identification technology
The importance of microorganisms to national biosafety has become increasingly prominent, and the demand for accurate identification and rapid detection of microorganisms is very urgent. Traditional microbial identification and detection methods are mostly dependent on the pure culture of microorganisms, thus making them time-consuming. In addition, they have low sensitivity and poor specificity. Furthermore, the automatic bacterial identification method is greatly affected by the concentration of the reaction substrate and the nutrient composition of the medium. The biochemical reactions, cell state, and metabolism of some bacteria make it difficult to achieve accurate identification. Therefore, microbial precision identification technology with high specificity and high sensitivity is widely needed in microbial health and safety industry. It is suggested that relevant scientific research and higher institutions are given policy and financial support to carry out in-depth research on molecular identification technology. On the basis of the existing microbial resources identification technology, there should be focus on the development of rapid and accurate identification technologies, strain identification research kit, and timely microbial identification strategies for classification and determination of function of complex microbial groups and for understanding their biodiversity and the relationship with ecological functions.
《4.2 Establishment of a microbial resource identification platform with independent intellectual property rights》
4.2 Establishment of a microbial resource identification platform with independent intellectual property rights
A DNA identification platform based on MNP marker technology should be established, which can detect 96 samples at a high throughput. More than 100 microbial targets can be detected for each sample, and each microorganism can be divided into subspecies or even races or variants, with an accuracy rate of 99.98 %. The localization of testing equipment, consumables, and analysis software can overcome the challenges of microbial resource identification. Moreover, it can solve the aerosol pollution problem of PCR technology and reduce the demand for laboratory physical space and high-level laboratories.
《4.3 Improving the database of microbial resources》
4.3 Improving the database of microbial resources
The establishment of the microbial resource database is of great significance to the microbial research in China. It is suggested that the focus should be on supporting the relevant scientific research units working on microbial resource identification technology, and policy and financial support should be given for the construction and improvement of three microbial resource databases, including National Microbial Resource Center, National Pathogen Resource Center, and National Virus Resource Center. Preserved microorganisms should be identified and database information should be improved. Moreover, there is need to provide support for microbial intellectual property protection and resource development and utilization as well as accelerate the industrialization process related to microbial health and safety. In addition, the construction of microbial genomics, metabolomics, transcriptomics, proteomics and other information resource databases should be accelerated to enrich the microbial resource database.














 京公网安备 11010502051620号
京公网安备 11010502051620号




