《1. Introduction》
1. Introduction
Haemophilus parasuis (H. parasuis ) is a nicotinamide adenine dinucleotide (NAD)-dependent Gram-negative bacterium that causes Glässer’s disease including pneumonia, arthritis, serofibrinous polyserositis, and meningitis in swine, leading to huge economic losses in the swine industry worldwide [1–3]. In the last few years, the number of outbreaks caused by H. parasuis has been reported to increase dramatically in Europe [4,5], Asia [6,7], and North America [8]. The high prevalence of non-typable strains and the serotype diversity of H. parasuis hinder the development of effective cross-protecting vaccines [2].
Antimicrobial treatment as an effective therapy continues to be indispensable for the control of Glässer’s disease. A number of antimicrobial agents, including β-lactams, aminoglycosides, macrolides, phenicols, tetracyclines (TETs), fluoroquinolones, and sulfonamides, have been used to control this disease through feed, water, or injection in swine. Antimicrobials at low dosages have also been used for growth promotion and disease prevention during swine production [9]. However, the extensive use of antimicrobials has been deemed the major cause of antimicrobial resistance (AMR) accumulation [10–12]. The AMR and AMR genes (ARGs) of H. parasuis in different geographical regions have been reported previously [13–17]. It has been reported that H. parasuis possesses β-lactams resistance (blaROB-1, blaTEM-1) [18], TETs resistance (tetB, tetC, tetH ) [17], sulfonamides resistance (sulI, sulII ) [19], and fluoroquinolones resistance (qnrA1, qnrB6, aac(6')-Ib-cr, gyrA, gyrB) [20].
Vietnam has the fifth-highest swine production in the world. However, there is no information about the prevalence of AMR phenotypes and ARGs or about their correlation, in H. parasuis isolates from Vietnam. In this study, we investigated the antimicrobial susceptibility profiles against 25 antimicrobial agents and detected the presence of ARGs in 56 strains of H. parasuis isolated from swine in the Quang Binh and Thua Thien Hue Provinces, Vietnam. The associations between different AMR phenotypes and genotypes were analyzed. These results provide a first glimpse of the prevalence and epidemiology of AMR in H. parasuis in Central Vietnam, which is useful for the clinical control of Glässer’s disease, as well as for the development of policies and clinical practice guidelines to reduce AMR in swine production in Vietnam.
《2. Materials and methods》
2. Materials and methods
《2.1. Bacterial isolates》
2.1. Bacterial isolates
A total of 56 H. parasuis strains were used in this study. The strains were isolated from swine in swine farms and slaughterhouses in Central Vietnam (Quang Binh Provinces and Thua Thien Hue) from June to September 2017, as characterized in our previous study [21]. The bacteria were cultured using tryptic soy agar (TSA; BD DifcoTM, BD Biosciences, USA) containing 10 μg·mL-1 of NAD (Sigma-Aldrich, Inc., USA) and 5% of bovine serum.
《2.2. Antimicrobial susceptibility testing》
2.2. Antimicrobial susceptibility testing
The broth microdilution method suggested by the Clinical and Laboratory Standards Institute (CLSI) was used to test the antimicrobial minimum inhibitory concentrations (MICs) of the H. parasuis strains [22]. A total of 25 antimicrobial agents, including gentamicin (GEN), kanamycin (KAN), streptomycin (STR), tobramycin (TOB), spectinomycin (SPE), amoxicillin (AMX), cephalexin (CFL), cefuroxime (CFX), penicillin (PEN), enrofloxacin (ERF), norfloxacin (NOR), ciprofloxacin (CIP), tiamulin (TIA), tylosin (TYL), erythromycin (ERY), lincomycin (LIN), chloramphenicol (CHL), florfenicol (FFC), sulfamethazine (SDM), trimethoprim/sulfametoxazole (TXT), doxycycline (DOX), TET, chlortetracycline (CTET), and colistin (CL), were used in the MIC test. The breakpoints for the antimicrobial agents were set according to the CLSI guidelines [22]. The ranges of resistance were recorded, along with the MIC50s and the MIC90s of the strains. Heomophilus influenzae ATCC 49247 was used as the quality-control strain. The strains that were resistant to at least two different types of antimicrobials were classified as multi-drug resistant (MDR).
《2.3. ARG amplification》
2.3. ARG amplification
Simplex polymerase chain reaction (PCR) assays were used to detect the presence of 32 ARGs that confer resistance to aminoglycosides (eight genes), β-lactams (two genes), fluoroquinolones (seven genes), macrolides (four genes), phenicols (five genes), sulfonamides (two genes), and TET (four genes). The ARGs, primers, and sizes of each amplified product are listed in Table 1 [17,19,23–29]. The PCR mixture had a total volume of 25 μL and included 12.5 μL of 2 × Taq PCR master mix (CW Biotech, China), 1 μL of each primer (25 μmol·L-1 ), 8.5 μL of ultrapure H2O, and 2 μL of genomic DNA (gDNA) for each strain (at > 10 ng·μL-1 ). All PCR assays were performed using an Eppendorf thermal cycler, with cycling conditions that were optimized for each target gene (Table 1). The PCR products were confirmed using 1% agarose gel electrophoresis and were visualized under ultraviolet light.
《Table 1》
Table 1 Primers used for ARG amplification.



F: forward; R: reverse.
《2.4. Data analysis》
2.4. Data analysis
Descriptive analysis was performed, and variables were recorded as necessary for statistical modeling using SPSS software (IBM SPSS Statistics version 18.0, IBM, USA). The primary outcome and response variables of interest included the individual resistance genes with prevalence rates of more than 2%. Strains with phenotype resistance to two or more antimicrobial agents were defined as multiple-AMR strains, and strains with two or more resistance genes in a single strain were defined as multiple-ARGs strains. The associations between specific ARGs and the AMR phenotype were calculated using Chi-square and Fisher’s extract tests. Statistically significant associations were shown as odds ratios (ORs) with 95% confidence intervals (CI). An OR of > 1 (a positive association) indicated the increasing probability of the cooccurrence of the genotype (or phenotype) being studied with the measured phenotype (or genotype), while an OR of < 1 (a negative association) indicated the decreasing probability of the co-occurrence of the genotype (or phenotype) being studied with the measured phenotype (or genotype). An association was significant if the p value was lower than 0.05.
《3. Results》
3. Results
《3.1. AMR phenotypes of H. parasuis strains》
3.1. AMR phenotypes of H. parasuis strains
The susceptibility of the 56 H. parasuis strains to the 25 antimicrobials is shown in Table 2. According to the MIC breakpoint of each antimicrobial agent, the strains showed high resistance to all of the antimicrobial agents except fluoroquinolones, TIA, DOX, and FFC. Among all of the strains tested, the highest resistance rate was observed for TXT (94.6%), followed by resistance to CL (91.1%), CHL (91.1%), PEN (85.7%), GEN (83.9%), LIN (82.1%), AMX (78.6%), CFL (71.4%), ERY (69.6%), CTET (67.9%), and TYL (66.1%). None of the H. parasuis strains were resistant to DOX. Moreover, the results revealed that all of the strains were MDR; in fact, a strain from nasal swab of swine in a slaughterhouse was surprisingly resistant to 18 antimicrobials (Table 3).
《Table 2》
Table 2 MICs for 25 antimicrobial agents of H. parasuis strains.

Breakpoints of resistance used are indicated with vertical black lines when available. Number of strains with MICs of trimethoprim/sulfamethoxazole (TXT) = 0.15/0.008, 0.30/0.016, 0.6/0.032, 1.2/0.064, 2.4/0.125, 4.8/0.25, and 9.5/0.5 are not available. MIC50, and MIC90: the lowest concentration of AM agents capable of inhibiting the growth of 50% and 90% of strains, respectively.
a MIC of TXT = 19 μg·mL-1/1 μg·mL-1.
b MIC of TXT = 38 μgmL-1/2 μgmL-1.
c MIC of TXT = 76 μgmL-1/4 μgmL-1.
d MIC of TXT = 152 μgmL-1/8 μgmL-1.
e MIC of TXT = 304 μgmL-1/16 μgmL-1.
f MIC of TXT = 608 μgmL-1/32 μgmL-1.
g MIC of TXT = 1216 μgmL-1/64 μgmL-1.
《Table 3》
Table 3 Numbers of AMRs and ARGs in H. parasuis strains.
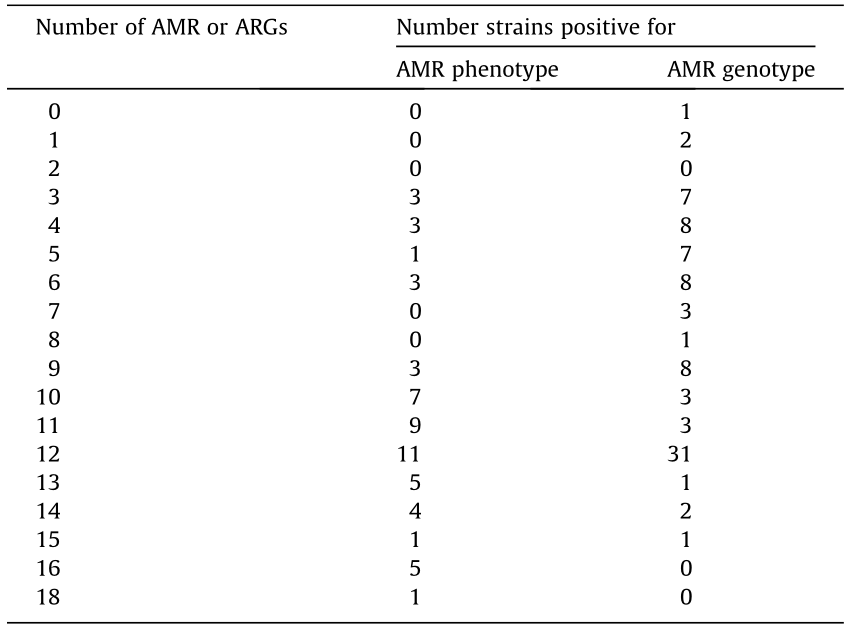
《3.2. Associations among AMR phenotypes》
3.2. Associations among AMR phenotypes
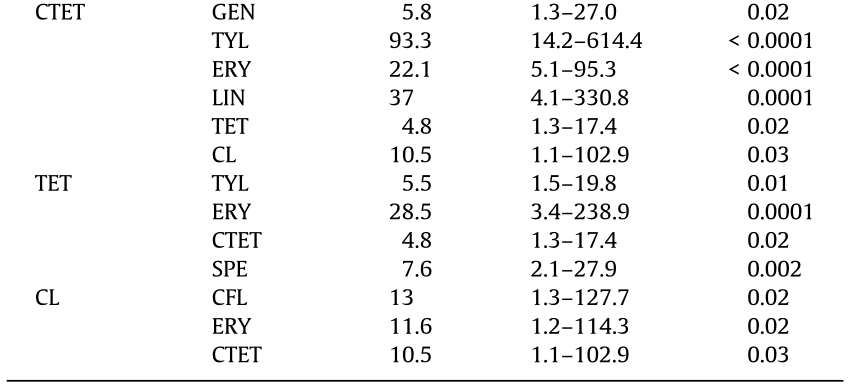
We next analyzed the relationship among the 16 AMR phenotypes. As shown in Table 4, almost every AMR was significantly associated with at least one other AMR (p < 0.05). The strongest association, which showed an increased probability of observing the outcome resistance in the presence of the predictor resistance, was found to be between TYL and CTET (OR = 93.3, p < 0.0001), followed by the association between TYL and ERY (OR = 65.6, p < 0.0001), and that between AMX and PEN (OR = 60.2, p < 0.0001). Strong associations between resistance to LIN and CTET as well as between resistance to CFL and PEN were also observed.
《Table 4》
Table 4 Outcome predictor association between AMR phenotypes of H. parasuis strains.


《3.3. Amrgenotypes of H. parasuis strains》
3.3. Amrgenotypes of H. parasuis strains
Table 5 presents the findings regarding the presence of each ARG in the 56 H. parasuis strains. The blaTEM-1 gene was the most frequently detected ARG (94.6%), followed by int (76.8%), gyrA (58.9%), and rmtD (50.0%). The parE and flor genes had the lowest detection rates (3.6% for both). However, none of the strains were positive for gyrB, cfr, or tetC. Most of the strains (55 strains, 98.2%) carried at least one of the ARGs (Table 3).
《Table 5》
Table 5 Distribution of ARGs in H. parasuis strains.


《3.4. Associations among resistance genes》
3.4. Associations among resistance genes
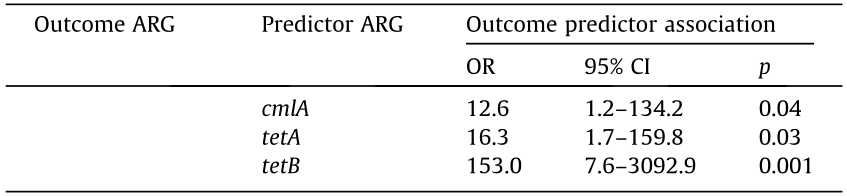
Positive associations (ordered by OR) were observed between the following gene pairs: tetB/tetH (OR = 153, p = 0.001), tetA/tetB (OR = 75, p = 0.001), tetH/aac(60 )-Ib (OR = 49, p = 0.001), sulII/aac (6')-Ib (OR = 23, p = 0.003), and tetB/aadA1 (OR = 23, p = 0.01) (Table 6). However, amtA, blaTEM-1, parE, and flor were not associated with any of the other genes. Most of the associations between ARGs were positive (OR > 1), but the associations between the four ARG pairs (qnrA1/gyrA, qnrB1/int, parC/int, and lnu(C)/int) were negative (OR < 1) (Table 6).
《Table 6》
Table 6 Pairwise statistical correlation between ARGs of H. parasuis strains.



Table 6 (continued)
《3.5. Associations between phenotypes and genotypes of antimicrobial resistance》
3.5. Associations between phenotypes and genotypes of antimicrobial resistance
Resistance genes can be linked to genetic elements, and the use of a particular antimicrobial can select for resistance not only to its own, but also potentially to a variety of other antimicrobials [30]. The strong association between the phenotypes and genotypes of AMR in bacteria supports the hypothesis that antimicrobial use selects for bacteria with novel resistance determinants [31]. Significant associations were found between AMR phenotypes and genotypes in the H. parasuis strains (Table 7). Among the 47 strains resistant to GEN, 33 strains (70.2%) carried one to seven corresponding ARGs, and this phenotype was strongly associated with the presence of rmtD (OR = 10.8, p = 0.02). For the 27 strains resistant to KAN, strA was detected in six strains (22.2%), strB in seven (25.9%), aadA in 16 (59.3%), amtA in one (3.7%), rmtB in 10 (37.0%), rmtD in 22 (81.5%), aacC2[aac(3)-lic] in seven (25.9%), and aadA1 in eight (29.6%). However, it is worth noting that one resistant strain did not carry any of the related resistance genes (3.7%). KAN resistance was associated with the presence of aadA (OR = 19.6, p < 0.0001), rmtD (OR = 16.9, p < 0.0001), aadA1 (OR = 11.8, p = 0.01), both strB and aacC2[aac(3)-lic] (OR = 9.8, p = 0.02), strA (OR = 8.0, p = 0.048), and rmtB (OR = 7.9, p = 0.01). Most strains (17/18, 94.4%) that were resistant to TOB contained the corresponding ARGs, including strB, aadA, rmtB, rmtD, and aadA1, among which strB (OR = 23.5, p = 0.001) and rmtB (OR = 11.7, p = 0.001) were found to have much stronger correlation than the others. Among the 18 strains resistant to SPE, strA was detected in six strains (13.3%), strB in eight (17.8%), aadA in 17 (37.8%), amtA in two (4.4%), rmtB in 11 (24.4%), and aadA1 in seven (15.6%). SPE resistance was associated with aadA (OR = 29.8, p < 0.0001), rmtB (OR = 22.5, p < 0.0001), and (rmtD) (OR = 9.6, p = 0.001). Thirteen strains were classified as CFX resistant, and all of these were found to carry blaTEM-1, while five (38.5%) also carried blaROB-1. In contrast, 40/53 (75.5%) strains carried blaTEM-1, and one out of six strains positive for blaROB-1 did not show phenotypes resistant to CFX. CFX resistance was strongly associated with blaROB-1 (OR = 26.3, p = 0.002). Twenty-five (67.6%) of the 37 TYL-resistant strains were positive for at least one corresponding TYL-resistance gene. Of these strains, six (16.2%) carried erm(A), 20 (54.1%) carried erm(B), 11 (29.7%) carried erm(C), and two (5.4%) carried lnu (C). TYL resistance was associated with erm(B) (OR = 10.0, p = 0.03), erm(C) (OR = 7.6, p = 0.04), and lnu(C) (OR = 0.1, p = 0.005). However, TYL resistance was only positively associated with two resistance genes (erm(B) and erm(C)). Regarding resistance to the phenicol group, 51 and five strains were found to be resistant to CHL and FFC, respectively. Forty-five (45/51, 88.2%) of the CHL -resistant strains carried at least one corresponding ARG. A positive association was observed between CHL resistance and int (OR = 18.7, p = 0.01), and FFC resistance and catl (OR = 25.1, p = 0.004) and cmlA (OR = 18.7, p = 0.01). However, other antimicrobials, including AMX, CFL, and PEN, were not associated with any corresponding resistance gene.
《Table 7》
Table 7 Associations between phenotypes and genotypes of AMR in H. parasuis strains.
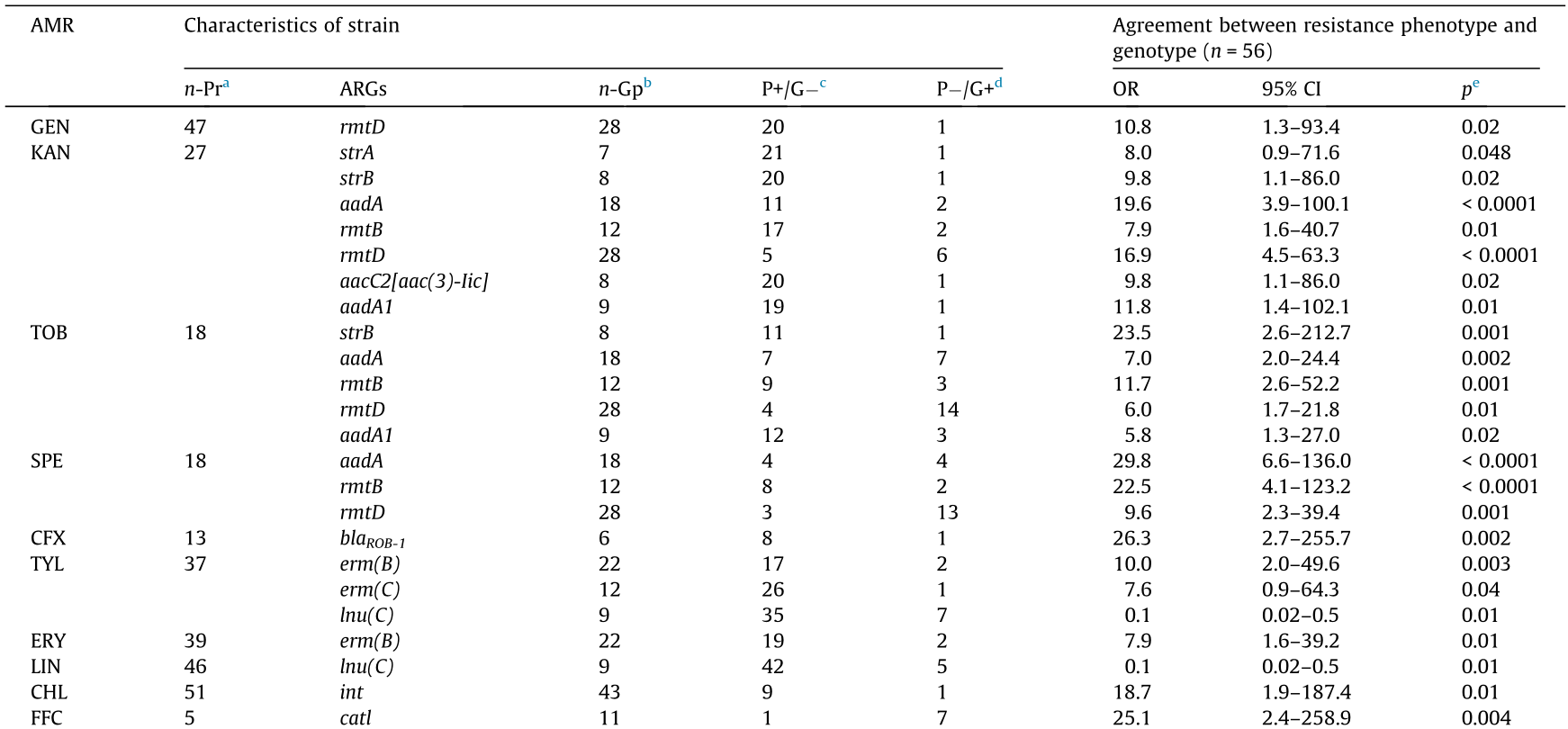

a n-Pr: number of strains expressing phenotype resistant to the indicated antimicrobial agents.
b n-Gp: number of strains carrying the indicated resistance genes.
c P+/G: number of phenotypically resistance strains (P+ ) with no resistance genes (G- ) for antimicrobials identified.
d P-/G+: number of phenotypically susceptible strains (P- ) with one or more resistance genes (G+ ) for antimicrobials identified.
e Only the results for AMR phenotype that displayed an association with those genotype at a p < 0.05 are shown.
《4. Discussion》
4. Discussion
In contrast to several other reports showing that most H. parasuis strains isolated from swine in the United Kingdom, Germany, and Denmark had lower levels of AMR [13,15,32], the data in this study revealed a high prevalence of AMR. Moreover, compared with the high-frequency resistance to both CIP and ERF that was found in the H. parasuis isolates from swine in Denmark [14] and the high prevalence of strains resistant to at least three antimicrobials (i.e., ERF, TXT, and CIP) in the strains (68.2%) from China [16] and Spain [15], our study suggests that the H. parasuis strains isolated from swine in Central Vietnam possess a lower rate of resistance to both CIP and ERF, as well as to at least three other antimicrobials. Our results are consistent with the results of a previous study by Nedbalcová and Kucˇerová [32], which showed that the H. parasuis strain isolated from swine in the Czech Republic possessed a low ratio of resistance to fluoroquinolones. Therefore, our findings suggest that fluoroquinolones, TIA, FFC, and DOX may be effective in controlling Glässer’s disease in swine produced in Central Vietnam. Moreover, the results of this study (Table 4) will be useful for antimicrobial users in practical applications.
Aminoglycoside resistance is conferred by the presence of amtA, rmtB, and rmtD encoding 16S rRNA-methyltransferase [33]; strA and strB encoding phosphotransferase [34,35]; and aadA, and addA1 encoding adenylyltransferase [36,37]. Our results showed that the H. parasuis strains carried strA, strB, aadA, and amtA genes. This is very similar to the results of a previous study [17], which reported that the H. parasuis isolates from swine in China carried rmtB (11.9%), rmtD (0.7%), aacC2[aac(3)-Iic] (4.2%) and aadA1 (20.8%), but does not align with other studies on the isolates from swine in Australia [25]. The highest detection rate of blaTEM-1 (94.5%) in the H. parasuis strains explained the high AMR phenotype to PEN (85.7%), AMX (78.6%), and CFL (71.4%). This resistance phenotype caused by the presence of blaTEM-1 and blaROB-1 (both encoding β-lactamases) has been reported previously [38–40]. The results of this study are similar to those reported for H. parasuis isolates from swine in China [18] and other countries [17,25,38], suggesting a wide spread of β-lactam resistance in H. parasuis. Previous studies have revealed that fluoroquinolone resistance is spreading and increasing rapidly in bacteria, because most qnr genes are located on a Tn-like sequence or integron on a conjugative plasmid [26,41]. In this study, although the frequency of strains resistant to fluoroquinolones was truly low, the bacterial strains carrying qnrA1 and qnrB1 could not be ignored. Furthermore, the macrolides resistance genes ermA, and ermB in H. parasuis have been reported to be at a low frequency in other studies [17,19]. However, our study revealed that the bacterial strains not only carried these genes but also carried other macrolides resistance genes such as ermC and lnuC. These genes are responsible for the ribosomal binding sites modification that is the most important macrolides resistance mechanism [42]. The emergence of FFC resistance in H. parasuis strains is attributable to a novel small plasmid pHPSF1 carrying flor [17], which explains the low frequency of the strains (in this study) carrying flor and FFC resistance capacity. Resistance to sulfonamide involves the presence of the sulI and sulII genes (encoding dihydropteroate synthases) associating with an integron system and a conjugative plasmid [43]. This is consistent with the results of this study, in which the two genes sulI and sulII (both sharing 8/56, 14.8%) were found in the H. parasuis strains. In addition, our investigation found that tetA, tetB, and tetH existed in the H. parasuis strains, which explains the phenotypic resistance to TETs, with the exception of DOX. Previous studies have shown that both tetB and tetH encoding the efflux pump are responsible for TET resistance in H. parasuis isolates from Australia [25,44]. The inactivation of TETs against the bacterial pathogen is involved in the presence of both tetB and tetH in the action of the efflux pump protein expelling the antimicrobial out of the cells.
Previous studies have demonstrated that the mechanisms of associations between resistance genes can be confirmed by molecular investigations [45,46]. An increased level of associations between resistance genes may result from the co-location of resistance genes on a single mobile genetic element such as a plasmid, transposon, or integron [43,47,48]. Our results showed many strong associations among ARGs which aligns with the opinion that there might be a linkage between many of these resistance genes on mobile elements. This opinion agrees with the results of a study by Rosengren et al. [31], which reported that qnrB1 had an increased association with eight other ARGs including rmtB, qnrA1, lnu(C), catl, int, tetA, tetB, and tetH. In addition, associations between sulI and lnu(C), and between sulII and tetA, are parts of integrons; the association between tetB and tetH is a requirement for the high frequency of strains resistant to CTET. The results strongly support the finding that the resistance genes are associated with a mobile DNA, such as plasmids and transposons, which enables horizontal gene spreading. Strong associations between the phenotypes and genotypes of AMR were found in our study, such as between GEN and rmtD, CFX and blaROB-1, and CHL and int, indicating that the resistance to a given antimicrobial was caused in some cases by a single gene. This finding is similar to the results of previous studies [30,31]. However, interestingly, we found that some strains possessed resistance phenotypes but did not have the corresponding ARGs and vice versa; for example, some strains showed resistance to AMX, but did not carry any corresponding genes. This finding is similar to the results reported by Rosengren et al. [31]. A possible explanation is that resistance phenotypes can be expressed upon the stimulation of many different genetic factors, and that each factor may present a unique epidemiological character [23,49]. Thus, the mechanisms of AMR in H. parasuis isolates from swine produced in Vietnam deserve further investigation.
《5. Conclusion》
5. Conclusion
This study is the first investigation on the prevalence of AMR and ARGs of H. parasuis isolates from swine in Central Vietnam. The strains tested were resistant to a broad range of antimicrobial agents with high MIC values, and high rates of multi-resistance were observed. The distribution of the most common resistance genes in the H. parasuis isolates included blaTEM-1, int, gyrA, and rmtD. This study also identified a number of ARGs that are clearly correlated with most of the AMR phenotypes observed in the H. parasuis strains. The results suggest that the use of some of the antimicrobials that show a high resistance frequency should be limited, while fluoroquinolones, TIA, FFC, and DOX can still be used in clinics in Vietnam to control Glässer’s disease. Moreover, the network of associations identified in this study will be useful for the development of policy and clinical practice guidelines to minimize AMR in Vietnam. It has been suggested that current attempts to limit the spread of AMR based on the prudent use of antimicrobials may prevent the selection of genes conferring resistance. Hence, assessment of AMR at the genetic level and the identification of associations between the phenotype and genotype resistance are critical tools in devising guidelines for the control of AMR.
《Acknowledgements》
Acknowledgements
This work was supported by the National Key Research & Development Program of China (2017YFD0500201), the Applied Basic Frontier Projects of Wuhan (2018020401011300), the Hubei Province Natural Science Foundation for Innovative Research Groups (2016CFA015), and the Fundamental Research Funds for Central Universities (2662018QD003).
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Chao Nguyen Van, Lijun Zhang, Tam Vu Thi Thanh, Hung Pham Hoang Son, Tuan Tran Ngoc, Qi Huang, and Rui Zhou declare that they have no conflict of interest or financial conflicts to disclose.














 京公网安备 11010502051620号
京公网安备 11010502051620号




