《1. Introduction》
1. Introduction
The spread of cancer cells from the primary tumor site to distant organs is the most devastating aspect of malignancy [1]. Pancreatic cancer is characterized as one of the most lethal and aggressive malignancies due to poor prognosis with remote and rapid metastatic spread [2]. Pancreatic ductal adenocarcinoma (PDAC) accounts for the vast majority (92%) of all pancreatic neoplasms, with an estimated five-year survival rate of less than 6% in clinical therapies [3,4]. The tendency of PDAC to metastasize, preferentially to the liver, results in high clinical lethality after surgical operation [5]. Recently, various studies have demonstrated that the hostile milieu of the liver is selectively preconditioned at an early stage to render it more conducive to the engraftment and growth of disseminated cancer cells [6], leading to a concept defined as pre-metastatic niche formation. Metastatic pancreatic tumor cells create intrinsic cellular properties that are selected for adaptation to specific organ environments [7]. An increasing number of studies on metastatic organotropism are uncovering crucial details regarding the manner in which pancreatic tumor cells metastasize to specific organs [8,9], revealing various key mediators that prepare metastatic niches in specific organs and identifying new targets that offer attractive options for therapeutic intervention [10]. Although many hypotheses have been presented, the theories of ‘‘seed and soil” and pre-metastatic niches have been particularly verified in pancreatic cancer progression [11]. However, the underlying mechanisms of organ-specific metastasis still remain to be elucidated.
Exosomes convey information by means of their vesicular contents and are considered to be a novel type of signaling mechanism between cells [12]. Tumor-derived exosomes contain abundant biological molecules secreted from the donor cells and function as signaling messengers for intercellular communication [10], contributing to the activation of various host cells, such as immune cells [13], cancer-associated fibroblasts (CAFs), and macrophages, that remodel the local stroma [14]. Notably, the modulation and reprogramming of these host cells by exosomes could create a metastatic preference environment for tumor cells in their target sites [15]. Tumor-derived exosomes may remodel the specific sites before the achievement of primary tumor cells by binding to the surface of target cells or fusing with the cell membrane [13,16]. For example, the integrin cargo within exosomes directs organspecific colonization by fusing with the host target cells in a tissue-specific manner, thereby initiating pre-metastatic niche formation [17]. PDAC-derived exosomes can support liver metastasis due to consequently increasing the liver metastatic burden in naive mice [18]. However, the precise mechanism by which tumorderived exosomes participate and influence the generation of a liver pre-metastatic niche is still poorly understood.
Cluster of differentiation 44 (CD44), a member of the transmembrane glycoprotein family [19,20], affects the activation of receptor tyrosine kinases (RTKs) [21]. In addition, it acts as a coreceptor for RTKs to mediate the signaling pathways associated with cell migration such as c-Met signaling [22]. Besides regulating various cellular, physiological, and pathological processes, CD44 serves as a receptor for extracellular matrix components [23], particularly hyaluronic acid (HA), to activate cytoskeleton proteins and thereby promote cell motility [24]. Many studies have demonstrated that CD44 is predominantly overexpressed in pancreatic cancer [25,26]; as both a cell–cell and a cell–extracellular matrix adhesion protein [27]. CD44 is positioned to contribute to cancer cells acquiring adhesion and migration properties [24,28]. Therefore, understanding the impact of CD44—particularly when it is delivered by exosomes during pancreatic cancer progression—could provide novel insights regarding the mechanism of tumor metastasis.
In the present study, we demonstrated that CD44 expression is positively correlated with pancreatic cancer prognosis. CD44 interacts with integrin α6b4 and promotes tumor cell proliferation, migration, and invasion by regulating the rat sarcoma (Ras) and extracellular signal-regulated kinase (ERK) signaling pathways. In addition, we found that pancreatic tumor-cell-derived exosomes can transfer CD44 to the target cell and cooperate with α6b4 in liver cells. Thus, exosomal CD44 generates a pre-metastatic milieu to facilitate specific organ metastasis by upregulating molecules such as CD133, α-smooth muscle actin (α-SMA), and interleukin6 (IL-6) in target liver cells. Exploration and identification of the effect of PDAC exosomal CD44 on liver metastasis could pave the way for developing diagnostic markers and therapies to halt metastasis.
《2. Materials and methods》
2. Materials and methods
《2.1. Patient sample collection》
2.1. Patient sample collection
Serum samples from patients with pancreatic cancer, with early-stage or late-stage pancreatic disease, were obtained from Ruijin Hospital affiliated to Shanghai Jiao Tong University School of Medicine, Shanghai, China.
《2.2. Cell culture》
2.2. Cell culture
The tumor cell lines PANC-1, Capan-1, PANC-1 CD44 knockdown (P-CD44kd), PANC-1 β4 knockdown (P-β4kd), Capan-1 CD44 knockdown (C-CD44kd), and Capan-1 β4 knockdown (C-β4kd) were cultured in Dulbecco’s Modified Eagle Medium (DMEM; Gibco, USA) with 10% fetal bovine serum (FBS; Gibco) and 1% penicillin/ streptomycin (P/S; Gibco). LX-2 cells were purchased from Life Technologies and cultured in Roswell Park Memorial Institute (RPMI) medium (Gibco) with 10% FBS. All cell lines were cultured in a humidified incubator containing 5% CO2 at 37 °C. For the in vitro culture of human LX-2 cells with various tumor-derived exosomes, cells were maintained in culture for 7 d, with media containing 500 μg·mL-1 of exosomes being replenished every other day.
《2.3. Establishment of knockdown cell lines》
2.3. Establishment of knockdown cell lines
Transfection and selection of short hairpin RNA (shRNA)-CD44 and shRNA-β4 clones were respectively performed. Vectorexpressed short hairpin RNAs were purchased from Stana Cruz Biotechnology (USA), and the oligonucleotide sequence of RNA-CD44 (sense: GACAGAAAGCCAAGTGGACTCAACGGAGA) and the HuSH shRNA green fluorescent protein (GFP) cloning vector (pGFP-V-RS-vector; Origene, USA) containing a non-effective shRNA cassette were used for the knockdown of CD44 expression. The oligonucleotide of RNA-β4 (sense: ATCGAAGCATTTCAAGA GAATGCTTCGATCATTACTTG) and pGFP-V-RS-vector containing a non-effective shRNA cassette were used for the knockdown of CD44 and β4 expression.
《2.4. Immunolabeling》
2.4. Immunolabeling
For exosome labeling, 1 mg of exosomes was labeled with 200 μL of fluorescent membrane tracer 3,3' -dioctadecyl-5,5' -di(4- sulfophenyl)-oxacarbocyanine, sodium salt (SP-DioC18; Invitrogen) in phosphate balanced solution (PBS, 1:10 000; Invitrogen) for 25 min. Labeled exosomes were washed twice and incubated with exosome-depleted fetal calf serum (FCS; Gibco) in the dark for 30 min so that the free dye could bind to proteins in the FCS. Dye-labeled exosomes were collected by centrifugation after washing with PBS. The labeled exosome pellets were purified using a 40% sucrose solution, as described previously. The dye-labeled exosomes were stored at –80 °C until further use. To stain cell membrane proteins, cells were fixed with 4% paraformaldehyde for 15 min and permeabilized with 0.5% TritonX-100 (Sigma, USA) for 20 min. Fixed cells were incubated with antiCD44, anti-α6β4, anti-α3β1, anti-matrix metalloproteinase (MMP)-9, anti-MMP-13, anti-epithelial cell adhesion molecule (EPCAM), anti-vimentin, anti-Src, and anti-Ezrin antibodies (Abcam, China) overnight at 4 °C and subsequently incubated with fluorescently conjugated Alexa Fluor 488 and Alexa Fluor 555 secondary antibodies (Thermo Fisher Scientific, USA).
《2.5. Western blotting analysis》
2.5. Western blotting analysis
Cells and exosomes were lysed with radioimmunoprecipitation assay (RIPA) buffer (Sigma) supplemented with protease and phosphatase inhibitors. Bicinchoninic acid (BCA) assay was performed to measure the sample protein concentration. The concentration of all the protein samples was corrected to 1 μg·μL-1 . The expression levels of CD44, α6β4, MMP-2, MMP-9, MMP-13, EPCAM, vimentin, Src, Ras, and ERK were assessed by Western blotting, and protein expression was normalized to β-actin. Protein samples (20 μg each) were resolved by sodium dodecyl sulfate– polyacrylamide gel electrophoresis (non-reducing). Protein samples were then transferred onto polyvinylidene fluoride (PVDF) membranes. The membranes were blocked with 5% fat-free dried milk solution at room temperature for 1 h and incubated overnight at 4 °C with anti-CD44, anti-a6b4, anti-MMP-2, anti-MMP-9, antiMMP-13, anti-EPCAM, anti-vimentin, anti-Src, anti-Ras, anti-ERK, and anti-β-actin antibodies (Abcam). Immunoblots were visualized by an enhanced chemiluminescence (ECL) detection system.
《2.6. Protein complex immunoprecipitation》
2.6. Protein complex immunoprecipitation
PANC-1, Capan-1, and P-CD44kd cells and exosomes were homogenized in lysis buffer (N-2-hydroxyethylpiperazine-N' -2- ethanesulfonic acid (HEPES) buffer (Sigma) with 1% Brij96 (Sigma)) for 60 min at 4 °C. Cell lysates (1 mg) or 100 μg of exosomal lysate were immunoprecipitated using the corresponding antibody (2 μg·mL-1 or 200 μL of hybridoma supernatant) overnight at 4 °C. Samples were precipitated with 5% protein-G sepharose (Invitrogen) for 1 h at 4 °C with rotation. Complexes were washed three times with lysis buffer. All liquid was removed using a 35 G needle attached to a vacuum pump to ensure minimal background. Complexes were dissolved in Laemmli buffer (Invitrogen), boiled at 95 °C for 5 min, and centrifuged briefly to separate the sepharose beads from the proteins.
《2.7. Exosome collection and isolation》
2.7. Exosome collection and isolation
For exosome collection, we first cultured PANC-1 cells in 10 cm dishes with DMEM supplemented with 10% FBS and 1% P/S antibiotic solution. When cells were covered 80% confluent of dishes, the culture medium was changed to DMEM without FBS for 48 h. The cell culture medium was collected and filtrated through 0.22 μm filters (Millipore, USA). This cycle was repeated two or three times. Exosomes were isolated by density gradient ultracentrifugation as follows: 500 × g (g: gravity speed) for 10 min twice to remove cells, 2000 × g for 20 min to remove residual debris, 10 000 × g for 120 min to remove micro-vesicles, and 100 000 × g for 2 h to collect nanovesicles in pellets. For exosome purification, sucrose gradient ultracentrifugation was performed and the 2–8 fractions of the solution were collected. Ultracentrifugation was performed with a high-speed Avanti JXN-30 centrifuge (Beckman Coulter, USA) and an optima JXN ultracentrifuge (Beckman Coulter).
《2.8. Clonogenic assay》
2.8. Clonogenic assay
PANC-1, P-CD44kd, and P-β4kd cells (200 and 1000 cells of each cell line) were seeded in a six-well plate (Corning, USA), and the number of colonies was counted after 10 d of culture. A clonogenic assay was performed to evaluate cell viability.
《2.9. Cell proliferation assay》
2.9. Cell proliferation assay
Cell growth was monitored using the cell-counting kit-8 (CCK-8) assay (Abcam, UK). In brief, PANC-1, P-CD44kd, and P-β4kd cells at a dilution of 5 × 103 cells per well were incubated in 96-well plates for 0, 24, 48, or 72 h, and CCK-8 buffer was added to the cells according to the manufacturer’s instructions. After 4 h of CCK-8 incubation, cell viability was measured by reading the optical density at a wavelength of 450 nm. All experiments were performed in triplicate.
《2.10. Cell cycle assay》
2.10. Cell cycle assay
A carboxyfluorescein succinimidyl ester (CFSE) staining assay was performed. 3 × 103 cells per well were collected and washed twice with sterile PBS. Cells were then labeled with 5 μmol·L–1 of CFSE and incubated for 24, 48, and 72 h at 37 °C. Thereafter, dilution of the CFSE-labeled cells was analyzed by flow cytometry.
《2.11. Cell apoptosis assay》
2.11. Cell apoptosis assay
Annexin V and propidium iodide (PI) labeling of cells were was performed to evaluate cell apoptosis. Fluorescein isothiocyanate (FITC) is used to staining cells for flow cytometry determination. PANC-1, P-CD44kd, and P-β4kd cells were seeded in a 96-well plate and incubated with cisplatin with a gradient concentration for 24, 48, and 72 h. Thereafter, cells were washed with PBS/1% FCS. Cells were incubated with Annexin-V-FITC/PI; cell apoptosis was then evaluated by flow cytometry using two channels: channel FL-1 for Annexin-V-FITC and channel FL-3 for PI.
《2.12. Cell migration assay》
2.12. Cell migration assay
The migration ability of PANC-1, Capan-1, and knockdown cell lines was tested in transwell chambers (6.5 mm; Costar, USA) with polycarbonate membranes (pore size: 8 mm). Cells were seeded in the upper chamber with 40 μL of DMEM. Five hundred microliters of DMEM with 10% FBS was added to the lower compartment. After 24 h incubation, cells that migrated to the lower surface of the filter membrane were stained with 0.1% crystal violet solution. Images were captured by light field microscope.
《2.13. Transmission electron microscopy》
2.13. Transmission electron microscopy
Exosome samples were collected by gradient ultracentrifugation and prepared for transmission electron microscopy (TEM) analysis. Exosomes were mounted on copper grids and fixed with 1% glutaraldehyde for 5 min in cold Dulbecco’s phosphate buffered saline (DPBS) to stabilize the immunoreaction. They were then washed in sterile distilled water, contrasted by uranyl-oxalate solution at pH 7 for 5 min. TEM was performed to capture pictures for exosome samples at a voltage of 80 kV.
《2.14. Flow cytometry》
2.14. Flow cytometry
Cell samples were detached using trypsin or ethylenediaminetetraacetic acid (EDTA; Sigma), then fixed and permeabilized in advance. Cells were incubated with the primary antibody (1–5 μg·mL-1 ) for 45 min; the fluorochrome-conjugated secondary antibody (0.3–0.5 μg·mL-1 ) was then added and the cells were incubated in the dark for 30 min at 4 °C. For intracellular staining, cells were fixed in 1% formalin for 20 min and permeabilized with 0.2% Tween in fluorescence activating cell sorter (FACS) buffer (Sigma) before incubation with the primary and secondary antibodies. Samples were acquired and analyzed using the FACSCalibur platform.
《2.15. Statistical analysis》
2.15. Statistical analysis
All data are expressed as the mean ± standard deviation (SD) of triplicate experiments and/or three repetitions. Statistical significance was determined using Student’s two-tailed t-test. Results with P < 0.05 were considered to be statistically significant.
《3. Results》
3. Results
《3.1. CD44 expression is positively correlated with tumor progression in pancreatic cancer》
3.1. CD44 expression is positively correlated with tumor progression in pancreatic cancer
CD44 is a multistructural and multifunctional cell surface molecule involved in various biological processes and is essential for the pathologic activities of cancer cells [26]. In order to explore the association of CD44 with pancreatic cancer metastasis, we first compared CD44 expression levels between normal individuals and pancreatic carcinoma patients in the XENA database↑ (↑ http://xena.ucsc.edu/ )and using the Oncomine browser (
( http://www.oncomine.org) . The results obtained from the XENA database indicated that CD44 expression in pancreatic cancer patients was higher than in normal individuals (Fig. 1(a)). To further determine the effect of CD44 in tumor metastasis, we also analyzed CD44 expression in different grades of patients with primary and metastatic tumors, respectively (Appendix A Fig. S1). Furthermore, the UALCAN browser↑ (↑ http://ualcan.path.uab.edu)was used to analyze the survival curve of patients from the cancer genome atlas (TCGA) database with high or low/medium CD44 expression levels. It was obvious that patients with high expression levels of CD44 had a poor prognosis compared with patients with low/medium expression levels of CD44 (Fig. 1(b)). Integrin receptors facilitate cell adhesion and mediate the activation of extracellular matrix with their large extracellular domains [29]. Thus, they bind to ligands via the extracellular domain, as well as the intracellular domain that interacts with cytoskeleton proteins [30]. Interestingly, we also found that the expression of CD44 in pancreatic cancer was positively correlated with α6β4, an integrin that binds to the cell membrane. We evaluated the expression levels of CD44 and α6β4 in blood serum samples collected from patients with early and late stages of pancreatic cancer. We found that the expression levels of both CD44 and α6β4 were higher in blood serum samples from patients with terminal stages of cancer than in samples from patients with early stages of cancer. It was notable that CD44 was positively correlated with α6β4 integrin in blood serum samples from patients with terminal stages of cancer, while the correlation between CD44 and α6β4 was not strong in blood serum samples from patients with early stages of cancer (Fig. 1(c)). These results from clinical samples were consistent with the analysis from the database. Therefore, CD44 could be a potential prognostic biomarker for pancreatic cancer and may be associated with the cell adhesion molecule α6β4.
http://www.oncomine.org) . The results obtained from the XENA database indicated that CD44 expression in pancreatic cancer patients was higher than in normal individuals (Fig. 1(a)). To further determine the effect of CD44 in tumor metastasis, we also analyzed CD44 expression in different grades of patients with primary and metastatic tumors, respectively (Appendix A Fig. S1). Furthermore, the UALCAN browser↑ (↑ http://ualcan.path.uab.edu)was used to analyze the survival curve of patients from the cancer genome atlas (TCGA) database with high or low/medium CD44 expression levels. It was obvious that patients with high expression levels of CD44 had a poor prognosis compared with patients with low/medium expression levels of CD44 (Fig. 1(b)). Integrin receptors facilitate cell adhesion and mediate the activation of extracellular matrix with their large extracellular domains [29]. Thus, they bind to ligands via the extracellular domain, as well as the intracellular domain that interacts with cytoskeleton proteins [30]. Interestingly, we also found that the expression of CD44 in pancreatic cancer was positively correlated with α6β4, an integrin that binds to the cell membrane. We evaluated the expression levels of CD44 and α6β4 in blood serum samples collected from patients with early and late stages of pancreatic cancer. We found that the expression levels of both CD44 and α6β4 were higher in blood serum samples from patients with terminal stages of cancer than in samples from patients with early stages of cancer. It was notable that CD44 was positively correlated with α6β4 integrin in blood serum samples from patients with terminal stages of cancer, while the correlation between CD44 and α6β4 was not strong in blood serum samples from patients with early stages of cancer (Fig. 1(c)). These results from clinical samples were consistent with the analysis from the database. Therefore, CD44 could be a potential prognostic biomarker for pancreatic cancer and may be associated with the cell adhesion molecule α6β4.
《Fig. 1》
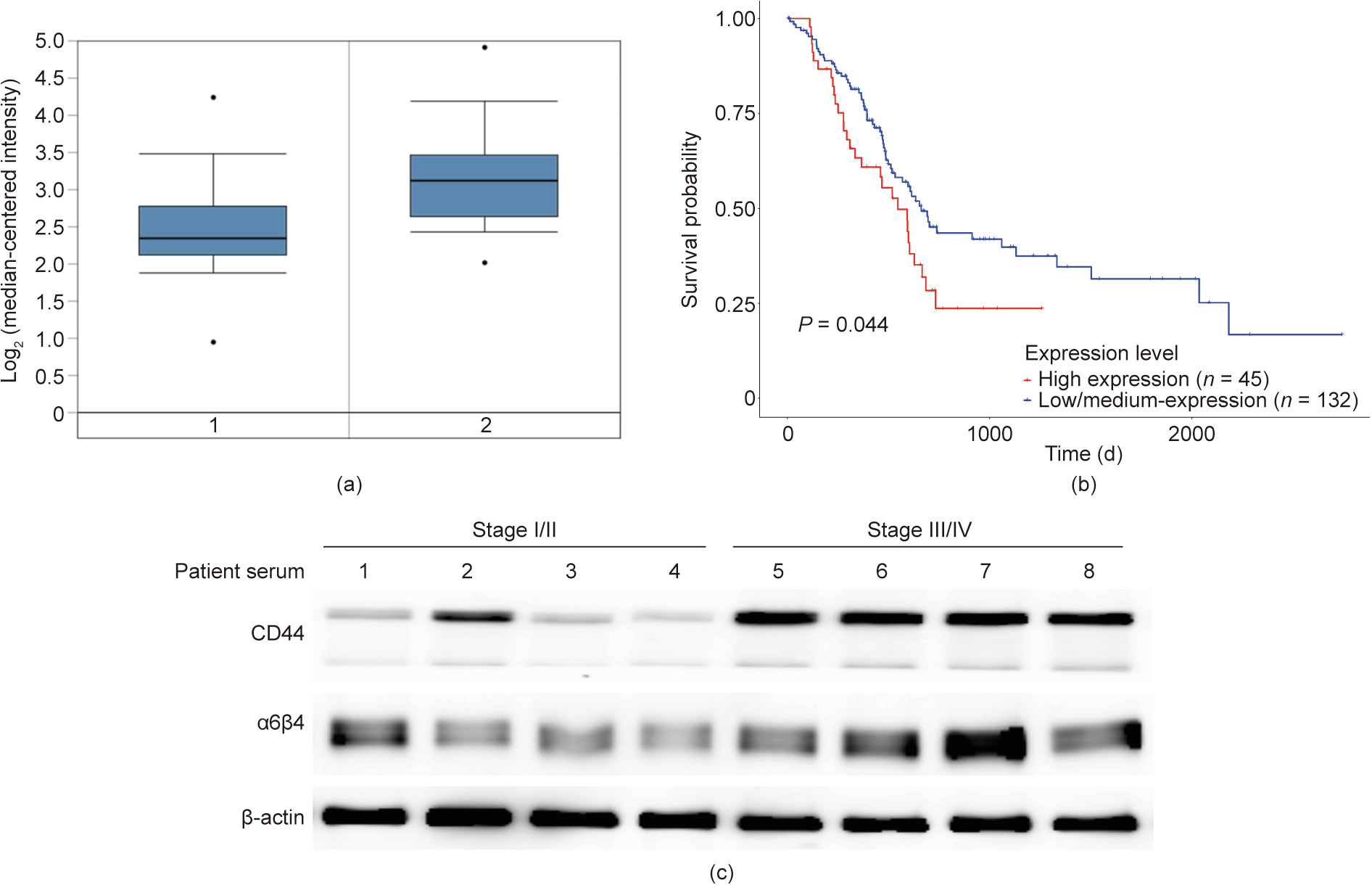
Fig. 1. CD44 is positively correlated with stage, tumor status, invasion, and poor prognosis in pancreatic cancer. (a) Differential expression of CD44 between normal and metastatic tissues of patient samples using the XENA dataset by mining the Oncomine database. Normal tissue is represented by 1 and tumor patient tissue is represented by 2, respectively. *: P < 0.05. (b) Kaplan–Meier survival curves display the overall survival rates of patients (data retrieved from the TCGA database and categorized based on CD44 expression levels using the UALCAN browser). (c) Western blotting was performed to detect the expression levels of CD44 and α6β4 in patient blood serum samples.
《3.2. CD44 regulates the expression of α6β4 in pancreatic cancer》
3.2. CD44 regulates the expression of α6β4 in pancreatic cancer
During the process of tumor metastasis, tumor cells usually gain enhanced migratory and invasive properties. Epithelial–mesenchymal transition (EMT) is also regarded as an initial sign of early tumor metastasis [16]. The CD44 molecule comprises an extracellular domain, which is expressed on the cell surface and is involved in cell motility [31]. Therefore, we explored the contribution of CD44 in pancreatic tumor cell migration and invasion. Two pancreatic cancer cell lines, PANC-1 and Capan-1, were used to examine the association between CD44 and other cell-motility-related genes. Considering the correlation between CD44 and α6β4 in patient samples, we first examined the expression levels of CD44 and α6β4 in these two cell lines. Both CD44 and a6b4 were highly expressed in these two cell lines (Fig. 2(a)). To further evaluate the effect of CD44 in tumor metastasis, CD44 knockdown (shRNA-CD44) cell lines were established using PANC-1 and Capan-1 cell lines, respectively. The expression level of α6β4 was downregulated upon inhibited expression of CD44 (Fig. 2(b)), suggesting that CD44 could directly regulate α6β4 expression. To explore the interaction between CD44 and α6β4, we performed a coimmunoprecipitation (Co-IP) experiment to confirm whether CD44 could form a complex with α6β4 in both the PANC-1 and Capan-1 cell lines (Fig. 2(c)). Furthermore, other cell-motility-associated genes, including Src, EPCAM, and Ras, were also found to be downregulated in the P-CD44kd and C-CD44kd cell lines, implying that CD44 may contribute to cell migration and invasion by regulating various signaling cascades (Fig. 2(d)). In addition, the inhibition of CD44 downregulated various proteases; as shown in the figure, MMP-9 decreased most significantly in the P-CD44kd and C-CD44kd cell lines, whereas the expression levels of MMP-2 and MMP-13 decreased by approximately 40% and 60%, respectively (Fig. 2(d)). Expression of Src, a protein-tyrosine kinase associated with cytoskeletal remodeling, also decreased in the P-CD44kd and C-CD44kd cell lines, suggesting that CD44 regulates Src expression to transform the structure of the cytoskeleton and contact positions between the cytoskeleton and the cell membrane, thereby promoting cell migration (Fig. 2(d)). Moreover, cell membrane staining was performed to evaluate the co-localization of CD44 and certain other EMT-associated proteins. The colocalization of CD44 and EMT-associated proteins evidently decreased in P-CD44kd, especially the co-localization of CD44 and α6β4. It was notable that CD44 was significantly co-localized with MMP-9, as well as vimentin and Ras, on the cell surface membrane of tumor cell lines; however, co-localization of these proteins decreased in the P-CD44kd and C-CD44kd cell lines. These results demonstrate that decreased expression of CD44 can downregulate the Src and Ras signaling pathways. In addition, reduced co-localization of proteins on the cell membrane may contribute to an early stage of the tumor metastatic process, leading to cell adhesion and migration (Fig. 2(e)).
《Fig. 2》
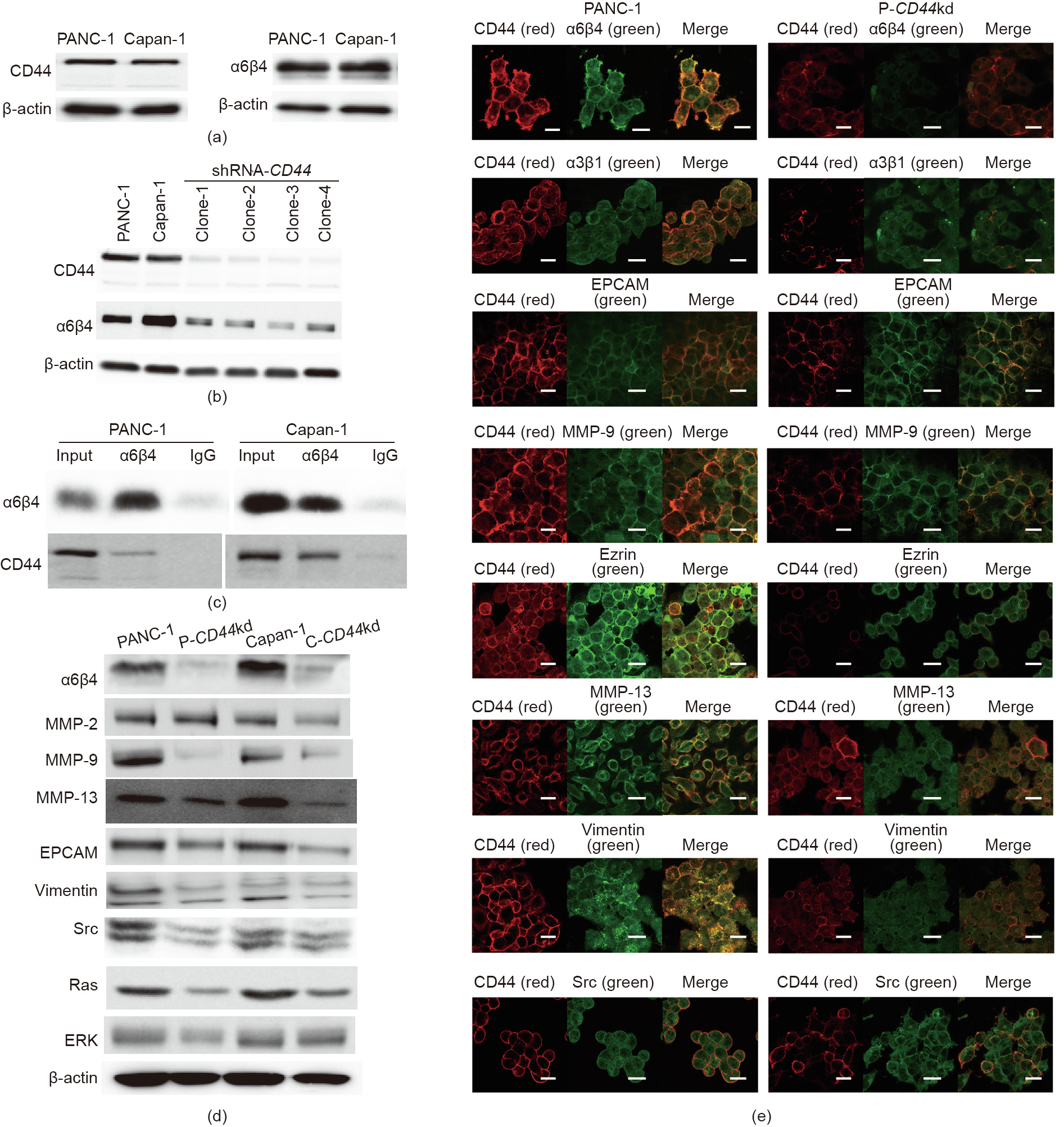
Fig. 2. CD44 regulates the expression of α6β4 in pancreatic cancer. (a) Two pancreatic cancer cell lines, PANC-1 and Capan-1, were used to examine the expression of CD44 and a6b4 by Western blotting. (b) shRNA-CD44 was used to knockdown CD44 in the PANC-1 and Capan-1 cell lines by silencing CD44 expression. (c) Co-IP of CD44 and α6β4. The results demonstrated that CD44 directly integrates with α6β4 in both the PANC-1 and Capan-1 cell lines, forming a CD44–α6β4 complex. Immunoglobulin G (IgG) is used as control. (d) Total protein was extracted and subjected to Western blot analysis to detect the expression of EMT-associated proteins in the PANC-1, P-CD44kd, Capan-1, and C-CD44kd cell lines. (e) Confocal microscopy determined the localization of CD44 and EMT-associated proteins and demonstrated the ability to assess the CD44–α6β4 complex on the cell surface. Images were obtained using a confocal microscope and were merged to assess protein localization (scale bar: 50 μm).
《3.3. CD44 affects cell proliferation and viability》
3.3. CD44 affects cell proliferation and viability
Abnormal cell proliferation is an important feature of tumor growth and aggressiveness. Therefore, we evaluated the effect of CD44 on tumor cell proliferation. To clarify the association between CD44 and β4, we established β4 knockdown (β4kd) cell lines using shRNA-β4. After shRNA-β4 was transfected into the PANC-1 and Capan-1 cell lines (P-β4kd and C-β4kd, respectively), a clonogenic assay was performed to evaluate pancreatic cancer cell viability. PANC-1, P-CD44kd, and P-β4kd were seeded (200 and 1000 cells of each cell line) in a six-well plate, and the number of cell colonies was counted after 10 d of culture. We observed that PANC-1 had the greatest number of colonies, while the number of colonies in the P-CD44kd and P-β4kd cell lines was around 30% and 50% less, respectively, compared with the PANC-1 cells (Fig. 3(a)). A CCK-8 assay was performed to monitor the cell growth in different cell lines, and the data showed little difference between the tumor and knockdown cell lines (Fig. 3(b)). Cell cycle analysis of the PANC-1, P-CD44kd, and P-β4kd cell lines was performed by CFSE labeling, and the results showed that the number of P-CD44kd and P-β4kd cells entering the third cycle after 72 h of culture was less than the number of tumor cells, suggesting that CD44 and β4 did not affect cell proliferation and cell cycling (Fig. 3(c)). Annexin-V-FITC/PI staining was performed to evaluate cell apoptosis, at 24, 48, and 72 h, in tumor cells with abnormal expression of CD44 and β4. The results showed that the number of apoptotic cells increased with increasing concentrations of cisplatin in the three cell lines. The apoptotic resistance of P-CD44kd and P-β4kd was reduced at all three tested time points compared with PANC-1 cells. The apoptotic rate of P-CD44kd and P-β4kd cells increased by approximately 10% compared with PANC-1 cells (Fig. 3(d)).
《Fig. 3》

Fig. 3. CD44 expression and cell proliferation and viability. (a) The impact of decreased CD44 and β4 on cell growth was measured by clonogenic assay. Cells (200 and 1000 of each cell line) were seeded in six-well plates; colonies were counted after 10 d of culture. *: P < 0.05. (b) The proliferation of tumor and knockdown cells was measured by CCK-8 assay. Cells were measured at the indicated time points and analyzed for proliferation ability using a CCK. OD: optical density. (c) Cells were labeled with CFSE to display the proportion of cells in different cell cycles, and were analyzed by FACS. (d) Cells were treated with gradient concentrations of cisplatin, and apoptosis was evaluated by Annexin-V-FITC/PI staining and cells were further stained with FITC to evaluated the fluorescent, the results were analyzed by FACS. *: P < 0.05.
《3.4. CD44–α6β4 complex affects cell migration and invasion》
3.4. CD44–α6β4 complex affects cell migration and invasion
Tumor cells achieve motility and invasion through changes in membrane structure and cellular actin; this leads to penetration of the surrounding tissues, resulting in tumor invasion and metastasis [32]. Thus, we determined cell adhesion and structure modulation in the PANC-1, Capan-1, P-CD44kd, C-CD44kd, P-β4kd, and C-β4kd cell lines. These cells were seeded into a 96-well plate; after 4 h of culture, cells were washed twice to remove non-adherent cells. Cell adhesion was evaluated by crystal violet staining of adherent cells. Enzyme-linked immunosorbent assay (ELISA) results showed that approximately 50% of the tumor cells adhered to the plate, whereas only approximately 30% of CD44kd cells were attached to the plate. Notably, only 10% of β4kd cells adhered to the plate. These results indicated that adhesion decreased in both CD44kd and β4kd cell lines (Fig. 4(a)), which implied that decreased expression of CD44 and β4 contributed to cells losing their capacity of connection and adhesion. To further investigate cell adhesion from a morphological perspective, a phalloidin staining assay was performed to determine the modulation of cell structure. Confocal microscopy was performed to observe cell antennae and, images were captured and analyzed. The confocal microscopy images revealed that the cell antennae of P-CD44kd, C-CD44kd, P-β4kd, and C-β4kd cells were reduced in comparison with tumor cells (Fig. 4(b)). Although we did not examine the association of CD44 and α6β4 with cell adhesion, we investigated the correlation of CD44 and α6β4 with cell migration in these cell lines.
《Fig. 4》
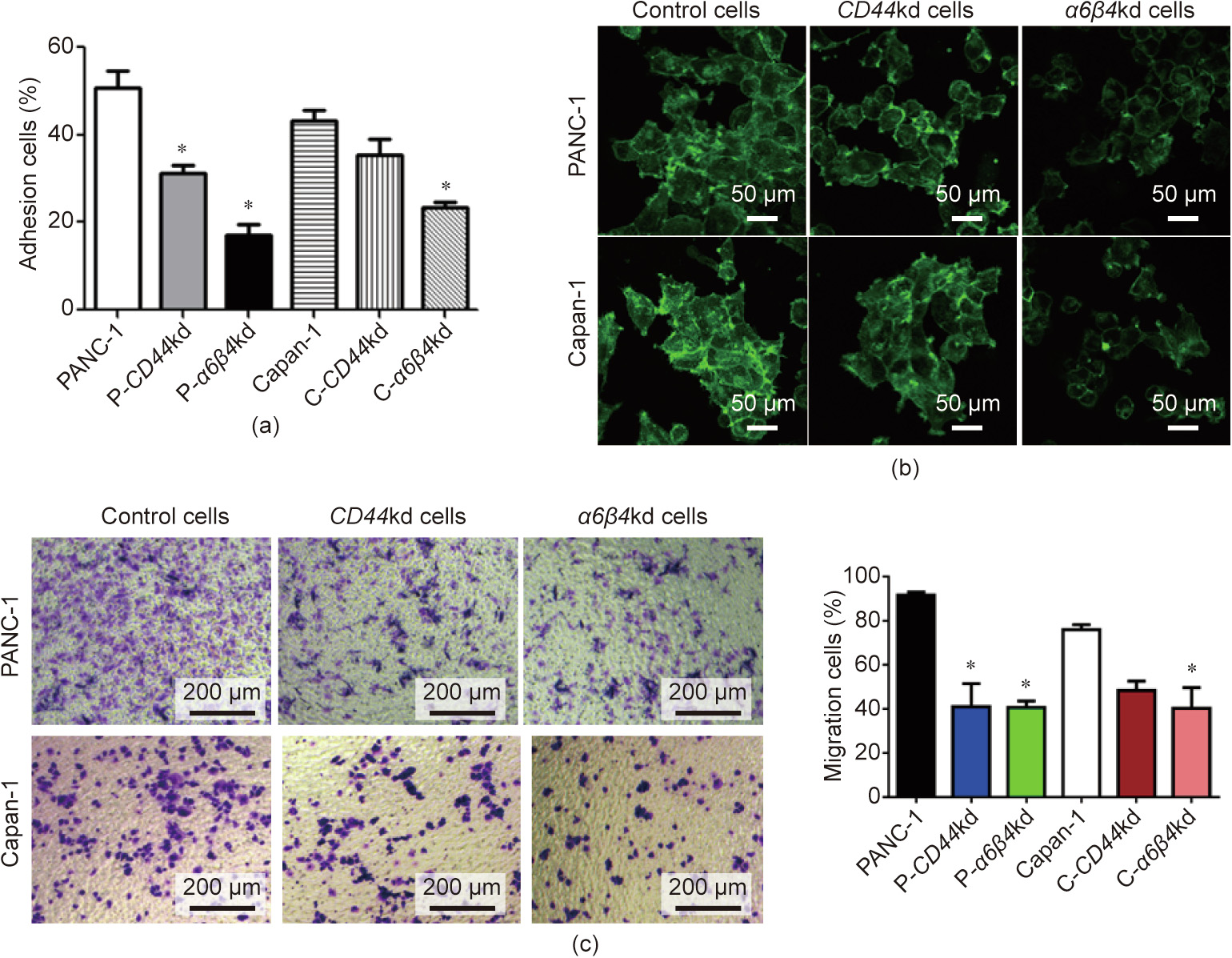
Fig. 4. CD44–α6β4 complex affects cell adhesion and invasion. (a) The impact of CD44 on tumor adhesion was evaluated by cell binding assay. Six cell lines were seeded in a 96-well plate for 4 h. Cells were washed and stained with crystal violet. The mean ± SD of the percent adherent cells is shown. *: P < 0.05. (b) The impact of CD44 on tumor antennae was evaluated. Cells were seeded on cover slips and stained with phalloidin fluorescein isothiocyanate. Confocal microscopy was performed to capture representative images. (c) The impact of CD44 on tumor migration was evaluated by transwell assay. Cells were seeded in the upper transwell chamber, and crystal violet staining of cells in the bottom wells displayed migration ability. *: P < 0.05.
Cell migration is a complicated process involving dramatic remodeling of the cytoskeleton and interaction between the cytoskeleton and the cell membrane [27]. CD44 can mediate cytoskeleton-associated proteins, which participate in the cytoskeletal remodeling process [22]. Thus, we next wanted to investigate whether CD44 and α6β4 also contribute to cell migration. Cells were seeded onto a transwell chamber, and the migrating cells were evaluated after 24 h. To analyze cell motility, cells were stained with crystal violet and observed under an optical microscope. The transwell assay results revealed that there were a reduced number of CD44–α6β4 knockdown (CD44–α6β4kd) cells migrating to the bottom wells, suggesting that CD44–α6β4kd weakened the migration ability of these cells (Fig. 4(c)). We also performed the matrigel invasion assay for cells, and found that the CD44-absent cells showed a significant decrease in tumor cell invasion (Appendix A Fig. S2). All these experiments indicated that the CD44–α6β4 complex impacted cell migration and invasion during the progression of tumor metastasis.
《3.5. CD44 cooperates with α6β4 in tumor-derived exosomes》
3.5. CD44 cooperates with α6β4 in tumor-derived exosomes
Exosomes transfer bioactive molecules from donor cells, which are taken up by neighboring tissues [33]; hence, they regulate target cell behavior. The PANC-1 and P-CD44kd exosomes were extracted by means of density gradient ultracentrifugation, and the morphology of the exosomes was examined using scanning electron microscopy. Both of PANC-1 and P-CD44kd exosomes showed cup-shaped nanostructures along with the similar size and quality (Fig. 5(a)). Nanoparticle tracking analysis (NTA) indicated the particle diameters of exosomes from tumor and CD44kd cells (Fig. 5(b)). Western blotting results showed that the expression level of CD44 in P-CD44kd exosomes was lower than that in PANC-1 exosomes, which indicated that P-CD44kd exosomes carried fewer CD44 membrane proteins from donor cells (Fig. 5(c)). These results demonstrated that tumor-derived exosomes contain similar proteins and their complexes in comparison with donor cells.
Considering that CD44 is enriched in exosomes due to endocytosis from the donor cell surface, we next evaluated the interaction between exosomal CD44 and α6β4 in exosomes. To verify whether CD44 and α6β4 might form a complex and be released into the extracellular space, a Co-IP assay was performed to assess tumorderived exosomes (Fig. 5(d)). Furthermore, the expression levels of integrin α6β4 in the PANC-1 and P-CD44kd cell lines were abolished using a combination of inhibitors such as Cilengitide and Cyclo (MCE, USA). Exosomes were isolated from these cells and pre-treated with inhibitors. We observed that the expression level of CD44 did not change significantly after blocking β4 expression in cells and exosomes, respectively (Fig. 5(e)). These results suggested that CD44 may not be regulated by integrin.
《Fig. 5》

Fig. 5. CD44 and α6β4 interacted with each other in exosomes. (a, b) Exosomes secreted from PANC-1 and P-CD44kd cells were isolated by ultracentrifugation and imaged by TEM; nanoparticle tracking analysis was performed to determine the size of the exosomes. (c) Immunoblotting analysis showed that the CD44 expression level in P-CD44kd exosomes was lower than the expression level in PANC-1 exosomes. (d) A Co-IP experiment was performed to examine the integration of CD44 and α6β4 in PANC-1 exosomes. IgG is used as control. (e) Immunoblotting analysis showing the expression of CD44 and α6β4 in PANC-1 cells and exosomes after pre-treatment with inhibitors. PANC-1 cells were pre-treated with inhibitors for 48 h to block integrin expression, and PANC-1-derived exosomes were isolated from the inhibitor-treated tumor cells. The CD44 in PANC-1 exosomes could interact with α6β4 to form a complex in vitro. Inh: inhibitor.
《3.6. Exosomal CD44 promotes metastasis via liver cell activation》
3.6. Exosomal CD44 promotes metastasis via liver cell activation
Exosomes can create a pre-metastatic niche in the target organ, thereby promoting metastasis of the target region [34,35]. Therefore, we determined the effect of CD44-competent exosomes on the uptake of target cells. Exosomes secreted from PANC-1 and P-CD44kd cells were stained with fluorescent dye diosp-18, and the dye-labeled exosomes were then incubated with the host cells LX-2, a human hepatic stellate cell (HSC) line. HSCs mostly transmit signals from sinusoid cells to the liver parenchyma, supporting the activation of proinflammatory cytokines and chemokines. To detect the contribution of exosomal CD44 in cell uptake and crosstalk, the PANC-1 tumor and P-CD44kd exosomes were incubated with LX-2 cells for 48 h. Interestingly, although the green fluorescent fields of both types of exosomes were captured at the same magnification, they showed no quantitative difference. Most of the PANC-1 exosomes entered into LX-2 cells, while P-CD44kd exosomes just attached to the surface of the target LX-2 cells and entered the extracellular space, indicating that the P-CD44kd exosomes lost the capacity for uptake (Fig. 6(a)). Next, we performed a transwell assay to evaluate the invasion ability of tumor exosomes modulated by liver cells. LX-2 cells were incubated with PANC-1 exosomes and P-CD44kd exosomes, and the migration rate of LX2 cells was detected. The exosome-modulated LX-2 cells were seeded onto the upper transwell chamber and, after 24 h of culture, migrating cells were stained with crystal violet (Fig. 6(b)). The microscopy images revealed that incubation with exosomes promoted the migration of cells to the bottom wells, and we observed an increased number of cells entering the bottom wells in the PANC-1 exosome incubation group compared with the P-CD44kd exosome incubation group.
Before pancreatic tumor cells metastasize to the liver, the expression of fibroblast activation protein (FAP) increases due to activated HSCs and myofibroblasts, which promotes the establishment of the pre-metastatic niche [36]. Overexpression of FAP increases the expression of CD44, suggesting cell adhesion and migration pathways mediated by CD44 in this epithelial cell line. FAP increases apoptosis, cell adhesion, and migration in the LX-2 human stellate cell line [37]. Thus, we evaluated the expression of a-SMA, FAP, and CD44 in control LX-2, PANC-1 exosomemodulated, and P-CD44kd exosome-modulated LX-2 cells, respectively (Fig. 6(c)). Western blotting analysis showed that the expression levels of cytokines and growth factors were upregulated in LX-2 cells, suggesting the activation of liver cells by pancreatic tumor exosomes. Notably, the CD44-competent exosomes displayed a stronger capacity to regulate molecules in the target LX-2 cells than CD44kd exosomes—particularly the expression levels of hepatocyte growth factor (HGF), α-SMA, hyaluronan (HA), and CD133 [38]—indicating that exosomal CD44 plays a critical role in the tumor metastatic process from the primary site of the tumor to the target organ (Fig. 6(c)). These results demonstrate that CD44 in exosomes has an influence on the invasive ability of target cells.
《Fig. 6》
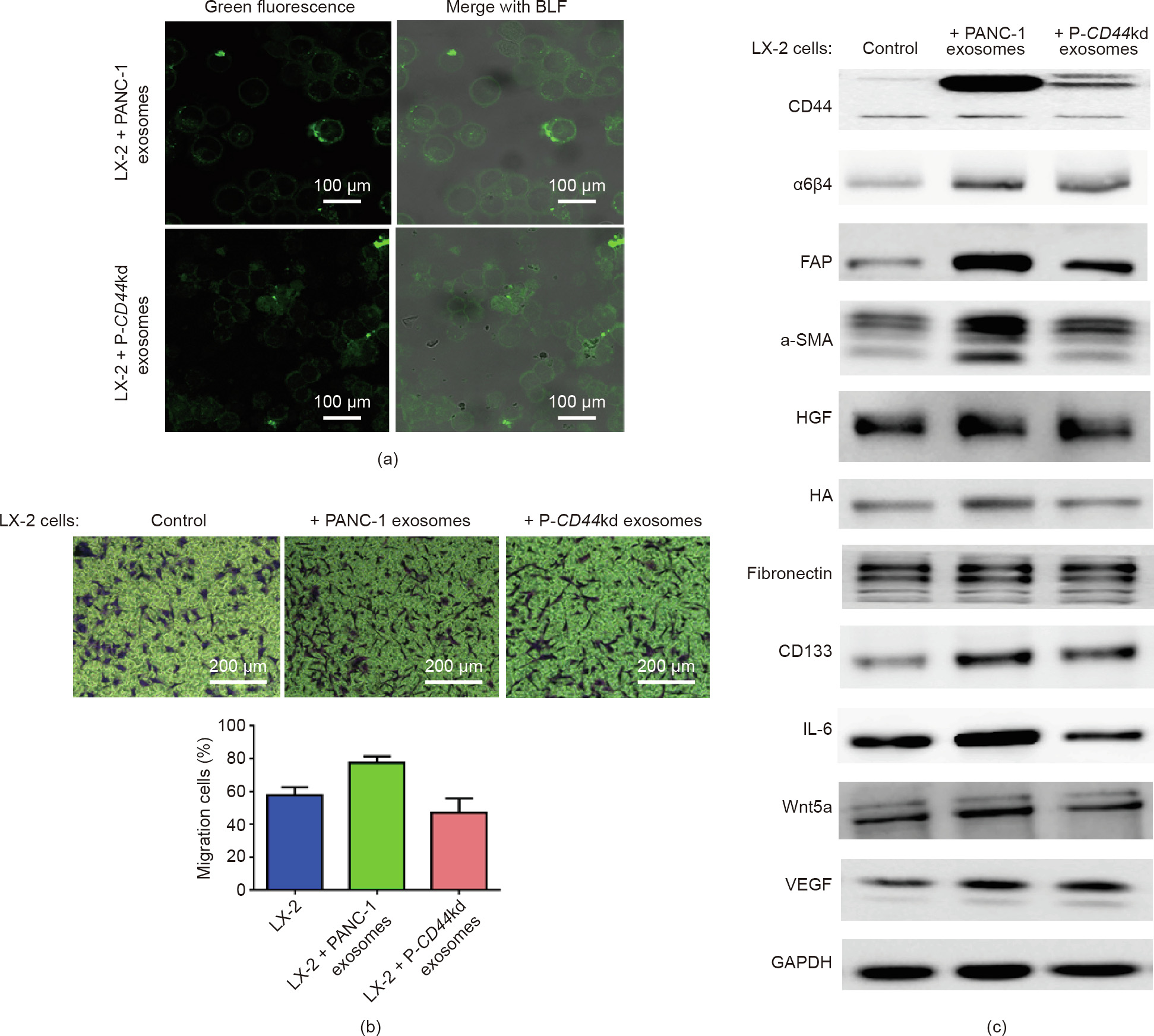
Fig. 6. CD44–α6β4 complex influences the migration and invasion of target cells. (a) Tumor-derived exosomes modulated target cells; 100 μg·mL-1 each of PANC-1 exosomes and P-CD44kd exosomes were co-cultured with LX-2 cells for 24 h. The dye-labeled exosomes, shown in the green fluorescence field, were secreted from the PANC-1 and P-CD44kd cell lines. Images were obtained by confocal microscopy. The green fluorescence was merged with a bright field (BLF), and the cells and exosomes were observed together. (b) The effect of exosomes on target cell migration was determined by transwell assay. LX-2 cells migrating to the bottom wells were stained with crystal violet. The number of cells was counted as an indicator to evaluate the invasion ability. LX-2 cells without exosome incubation were regarded as blank control. (c) The expression levels of cytokines and growth factors in the target cells were evaluated. Cells were pre-treated with tumor-derived exosomes. LX-2 cells were incubated with PANC-1 and CD44kd exosomes and the expression levels of the indicated proteins were evaluated. LX-2 cells without exosome incubation were regarded as the blank control group. β-Actin was used as a protein loading control. HA: hyaluronan; VEGF: vascular endothelial growth factor; GAPDH: glyceraldehyde-3-phosphate dehydrogenase.
《4. Discussion》
4. Discussion
Pancreatic cancer is an aggressive malignancy with a five-year mortality of 97%–98%, usually due to widespread metastatic disease [7,39]. Approximately 60%–70% of pancreatic cancer patients have peritoneal metastasis at the time of surgery. Thus, there is an urgent need to identify sensitive and specific biomarkers for the early detection and prognosis of pancreatic cancer [40]. CD44 has been reported as a potential biomarker correlated with tumor progression and poor prognosis in various cancers. Interestingly, CD44 expression was found to be enriched at the tumor edge of metastatic tumor specimens compared with the tumor center, which emphasized the importance of CD44 in metastasis [41]. Despite recent studies showing that a high level of CD44 expression is linked to several types of cancers, including pancreatic cancer, the mechanisms by which CD44 functions in pancreatic cancer metastasis and whether integrin α6β4 interacts with CD44 and acts as a mediator are still largely unknown. In this study, we verified the positive correlation between high expression levels of CD44 and pancreatic cancer incidence and prognosis. Furthermore, using clinical samples, we found that the expression levels of α6β4 were positively linked to the stage of pancreatic cancer. In addition, our experiments demonstrated that CD44 can integrate with α6β4 to form a complex, which can regulate signaling pathways such as the Src and ERK pathways. Meanwhile, focal adhesion kinase (FAK) is a non RTK that is critical for integrin-mediated signaling and cellular functions. FAK also functions as a convergent point for various signaling pathways particularly nuclear factor (NF)-κB that are associated with tumor motility and oncogenic transformation. The combination of CD44 and α6β4 also promotes tumor cell migration, invasion, proliferation, and apoptosis. For the first time, we found that CD44 carried by pancreatic-cancer-derived exosomes can interact with α6β4, thereby modulating target cell motility. The exosomal CD44–α6β4 complex functioned not only on pancreatic cancer cell membranes, but also during the uptake of exosomes by neighboring cells (Fig. 7). It is well implied that CD44 is not only a potential biomarker, but could also be applied in targeted therapy.
《Fig. 7》
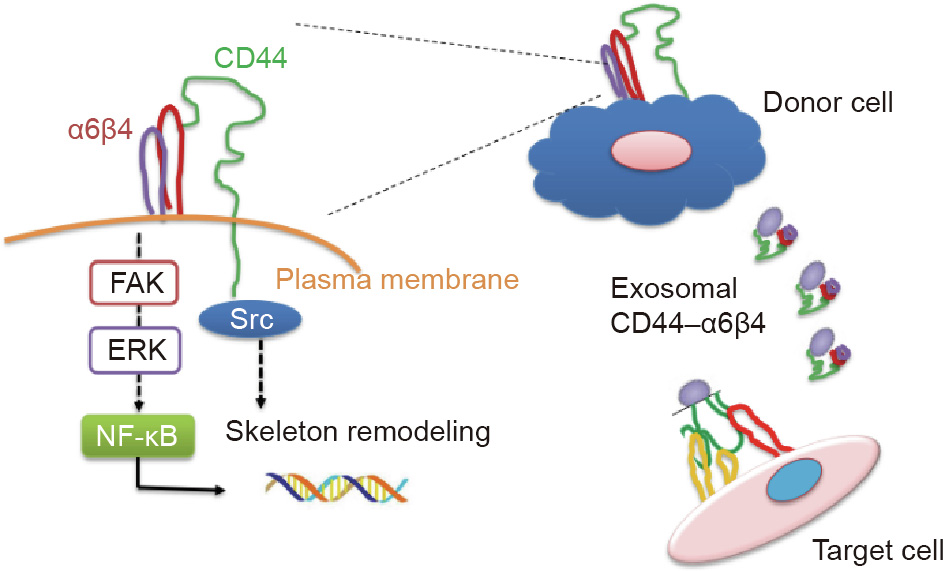
Fig. 7. Schematic model illustrating the regulation and effect of CD44 and integrin in pancreatic cancer liver metastasis.
Metastasis to vital organs such as bone, lung, liver, and brain is responsible for the vast majority of PDAC deaths [42]. It has been clearly recognized that metastatic seeding and growth are determined by both the intrinsic genetic properties of tumor cells and the local characteristics of the stromal microenvironment [43]. Understanding the organ-specific functions of metastasis-related signaling pathways may provide new opportunities for therapeutically targeting metastasis in different organs. Studying the liver metastasis of human pancreatic cancer is complicated due to several difficulties. In fact, liver metastatic lesions cannot be obtained for detailed analysis since liver surgery is not a treatment option for almost all patients with metastatic pancreatic cancer [8,44]. Thus, minimal functional genomic analysis of in vivo selected high metastatic variants of pancreatic cancer cell lines have led to the identification of distinct sets of organ-specific (e.g., bone, lung, and brain) metastatic genes [45]. HSCs actively participate in establishing the cancer pre-metastatic niche and are reprogrammed by cancer cells to facilitate tumor growth, becoming cancer-associated HSCs. In the present study, we focused on one of the upregulated genes in lymph-homing tumors, CD44 [14,20]. CD44 is a transmembrane protein that has previously been implicated in the homing of normal lymphocytes to the nodes. CD44 can induce many activities associated with the tumor metastatic process, such as uncontrolled cell growth, cytoskeletal reorganization, and intensive cell migration [46]. For example, the CD44–Ras pathway is involved in the activation of cell proliferation, the CD44– Rho family pathways are associated with cell migration, and the CD44–Akt pathway is linked to cell survival [31]. Recent studies have also reported that CD44 can interact with different types of membrane proteins to form complexes and hence promote the malignant cascade. Defined expression of the adhesion molecules and their corresponding ligands in host organs and on tumor cells govern organ and selective non-random tumor arrest [47]. For example, E-cadherin-integrin crosstalk regulates the dynamic interplay between epithelial cell–cell adhesion and cell–matrix adhesion signaling and contributes to the plasticity of tumor cells, driving effective migration and invasion. CD44 and integrins can mediate the expression of similar proteins, such as the Rho family and Src [30]. CD44 has been demonstrated to be the critical coreceptor of integrin αvβ3 to mediate the phosphorylation of Runx2 and Smad5, and to be a receptor activator for NF-κB ligand signaling during prostate cancer metastasis. Thus, CD44 determines the association between integrins and other receptors [48]. Therefore, it was necessary to explore whether CD44 and integrins could interact with each other to enhance their functions in tumor metastasis. Herein, our study reveals that CD44 can interact with integrin α6β4 and form a CD44–α6β4 complex.
Exosomes are cell-derived micro-vesicles enriched in particular proteins, lipids, mRNAs, and miRNAs of the donor cells [43], serving as intercellular communicators in a systematic way [11]. Several studies suggest that tumor exosomal molecues such as integrins can determine organotropic metastasis by fusing with organ-specific resident cells to establish the pre-metastatic niche [49,50], which in turn display various affinities for secondary targets, along with non-random patterns of dissemination. Exploring the effect of exosomal molecules on target organ metastasis could pave the way for developing diagnostic markers and therapies to halt metastatic spread. Previous studies have reported that CD44, as a transmembrane protein, is expressed in the exosomes secreted by many types of cancer cells, including breast cancer, ovarian cancer, and pancreatic cancer [51]. Exosomal CD44 can activate MMP9, a type of metalloproteinase, and thereby facilitate degradation of the extracellular matrix to promote cell motility [14]. Thus, it is essential to explore the mechanism by which CD44 participates in exosome delivery. CD44 is directly involved in extracellular matrix remodeling and serves as a transmembrane receptor for many extracellular matrix components, such as HA [51]. The combination of CD44 and HA can not only promote tumor cell arrest and extravasation, but also modify the surrounding matrix to support cell colonization, thus promoting cell proliferation [52]. Our study showed that exosomal CD44 can not only activate matrix proteins, but also interact with integrin α6β4 to form a complex and mediate downstream signaling pathways [47], such as the cSrc and the Ras cascades, to influence cell migration.
In summary, we demonstrated the connection between the CD44–α6β4 complex and the tumor metastatic process from two perspectives. First, the CD44–α6β4 complex can directly mediate cytoskeleton-associated proteins to activate downstream signaling pathways and promote pancreatic cancer metastasis. Second, exosome delivery indicates that pancreatic-cancer-derived exosomal CD44 can integrate with α6β4 on the recipient cells to transmit information associated with tumor invasion. Thus, our study provides new insight into the role of CD44 and the associating integrin in the tumor metastatic process. Furthermore, CD44 could be a novel prognostic marker and potential therapeutic target for pancreatic cancer.
《Acknowledgments》
Acknowledgments
This work was sponsored by grants from the National Natural Science Foundation of China (81803269 and 81427805), the Science and Technology Commission of Shanghai Municipality (18YF1412100 and 2019Y0150), the National Key Research and Development Program of China (2018YFC2000700), the Key Research Program of the Chinese Academy of Sciences (ZDRW-ZS-2017-1), and Shanghai Municipality Health Commission (GWV-10.2-YQ17 and 2019Y0150).
《Authors’ contribution》
Authors’ contribution
Wei Mu, Yang Ge, and Wenbo Wang performed and analyzed experiments; Pengfei Gu and Yajie Xu performed the experiments and data analysis. Wei Mu and Hui Wang wrote the manuscript, and Yang Ge corrected the manuscript draft. All authors approved the manuscript. All authors approved the manuscript.
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Wei Mu, Yajie Xu, Pengfei Gu, Wenbo Wang, Jingquan Li, Yang Ge, and Hui Wang declare that they have no conflict of interest or financial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2020.08.013.













 京公网安备 11010502051620号
京公网安备 11010502051620号




