《1. Introduction》
1. Introduction
Great success has been achieved in antiviral research with the chemotherapy of viral diseases induced by human immunodeficiency virus (HIV), hepatitis C virus (HCV), and hepatitis B virus (HBV). However, the outbreaks of influenza A virus (IAV), severe acute respiratory syndrome coronavirus (SARS-CoV), Middle East respiratory syndrome coronavirus (MERS-CoV) and, recently, SARS-CoV-2, have indicated the urgent need for drugs that are effective against respiratory RNA viruses. Infections by these viruses are often characterized by flu-like symptoms. They are highly contagious via aerosol transmission, spread in the asymptomatic stage, and have a long incubation period before the onset of symptoms. These viruses have caused pandemics with high mortality rates, especially in older adults and those with pre-existing medical conditions. As of 30 April 2021, coronavirus disease 2019 (COVID-19) has caused approximately 151 million infections and more than 3.18 million deaths worldwide [1]. The discovery of novel antiviral agents against respiratory RNA viruses is a priority, but it is a formidable task.
Although some antiviral drugs have been developed through rational design followed by chemical synthesis, the original discoveries are often inspired by natural products. During evolution, many microorganisms have developed diverse mechanisms to combat viruses and bacteria. Secondary metabolites of microorganisms are a source for potential antiviral candidates [2–4]. About 10% of the known 16 500 bioactive secondary microbial metabolites demonstrate antiviral activity [5]. For example, ribavirin (RBV), a nucleoside analogue for multiple antiviral treatment regimens such as those for IAV, HCV, dengue virus, and norovirus, was derived from pyrazomycin, which was isolated from Streptomyces candidus NRRL 3601 [3,4,6]. Vidarabine, the first antiviral drug approved by the US Food and Drug Administration (FDA) for antiviral treatment against systemic herpes simplex virus, was isolated from the marine sponge Tethya crypta [4,7]. Aristeromycin, a broad-spectrum antiviral compound, was isolated from Streptomyces citricolor in 1967 [8,9]. Other compounds with antiviral activity, such as formycin, myriocin, and cyclosporine A, are also from microbial sources [3,4]. Understanding the biosynthesis mechanism of antiviral antibiotics that can inhibit virus replication might aid in the discovery of novel antiviral drugs.
Streptomyces sp. 1647 was isolated from soil samples from southern China. Its fermentation broth or extracts have exhibited excellent anti-IAV activity since the 1970s. However, its antiviral chemical components remained a mystery until recently. In this study, we describe a multi-omics-based workflow that involves genomics, transcriptomics, and metabolomics analyses for determining the antiviral compounds, initiated by identifying the biosynthetic gene cluster (BGC) responsible for their production in the genome of strain 1647. This is an efficient approach for discovering novel antiviral candidates produced by microorganisms, in comparison with the traditional active natural product discovery process. A series of novel pseudo-tetrapeptides with excellent antiIAV and anti-coronavirus activities were identified. These agents provide potential lead compounds for developing new antiviral drugs.
《2. Materials and methods》
2. Materials and methods
《2.1. Strains, plasmids, and fermentation culture of Streptomyces sp. 1647》
2.1. Strains, plasmids, and fermentation culture of Streptomyces sp. 1647
All of the strains and plasmids used in this study are listed in Table S1 in Appendix A. Streptomyces sp. 1647 (China Pharmaceutical Culture Collection No. CPCC 200451) and its derivatives were grown at 28 °C on solid yeast malt glucose (YMG) medium (yeast extract 1.0%, malt extract 1.0%, glucose 1.0%, agar 1.5%; pH 7.2) for growth and seed culture. Tryptic soy broth (TSB) medium (BD, USA) was used for the isolation of the total DNA. A1, A2, and A3 media were mainly used as the fermentation media. The detailed components of these fermentation media are given in the Methods part in Appendix A. Escherichia coli (E. coli) DH5α was used as the general cloning host, and E. coli ET12567/ pUZ8002 was applied for the conjugative transfer according to the established protocol [10].
《2.2. Construction of strains for overexpression or knockout of the corresponding gene》
2.2. Construction of strains for overexpression or knockout of the corresponding gene
The primers used are listed in Table S2 in Appendix A. To obtain the recombinant plasmids, the target gene was inserted into vector pL646 [11], a pSET152 [12] derivative plasmid containing ΦC31 phage attachment site and integrase gene (attP-int) and a strong constitutive promoter ermE*p, through double enzymatic digestion and ligation for gene overexpression. The vector pOJ260 [12], a nonreplicating vector in Streptomyces, was used for gene knockout in Streptomyces sp. 1647. To inactivate the biosynthetic gene, two upstream and downstream homologous fragments of about 2.0 kilobases (kb) were inserted in the vector in order to generate a recombinant plasmid. The recombinant plasmid was introduced into Streptomyces sp. 1647 by conjugation. Then, the single exchange mutants were screened on a mannitol soya flour agar medium [10] containing apramycin and confirmed by polymerase chain reaction (PCR) analysis. After subculture for five generations on mannitol soya flour medium plates without apramycin, the spores were collected and diluted to isolate the apramycinsensitive mutants. The correct knockout mutants were confirmed by PCR, sequencing, and quantitative reverse transcription PCR (qRT-PCR).
《2.3. Bioactivity-based molecular networking》
2.3. Bioactivity-based molecular networking
The fermentation supernatants of Streptomyces sp. 1647 and its derivatives were concentrated five times using a Sep-Pak C18 solid-phase extraction column (Waters, USA). Liquid chromatography–mass spectrometry (LC–MS) data of the above samples were collected in negative full-scan mode with an Agilent 6510 quadrupole time-of-flight (Q-TOF) LC–MS instrument with a mass scan range set from 300 to 2000 m/z using the following conditions: column, Capcell-Pak MG II C18 column (150 mm × 4.6 mm, 5 μm); flow rate, 1.0 mL∙min–1 ; solvent A, acetonitrile; solvent B, 0.1% formic acid (v/v) in water; linear gradient of 5%–30% A/B (v/v) for 30 min followed by 30%–95% A/B (v/v) for 30 min; column temperature, 40 °C. The liquid chromatography–tandem mass spectrometry (LC–MS/MS) data were analyzed using the Global Natural Products Social (GNPS) molecular networking database↑ and the results were visualized and analyzed using Cytoscape software [13].
↑ http://gnps.ucsd.edu
《2.4. Isolation and identification of active components from Streptomyces sp. 1647》
2.4. Isolation and identification of active components from Streptomyces sp. 1647
The supernatant of the fermentation broth of the Streptomyces sp. 1647 strain was collected and adsorbed by macroporous adsorption resin (Diaion HP20; Mitsubishi, Japan). The column was then rinsed with twice the column volume of deionized water. Gradient elution was carried out with 20%, 50%, and 100% ethanol–water (v/v), respectively. Each gradient was eluted until the effluent had no color and designated as crude extracts 20E, 50E, and 100E, respectively. The eluates of each gradient were separately collected, condensed by evaporation, and lyophilized. Octadecylsilyl silica gel (ODS-A-HG; YMC, Japan) was used for the open column chromatography to conduct further separation of the lyophilized bioactive collections. High-resolution electrospray ionization mass spectrometry (HR-ESI-MS) was carried out on an Agilent G6500 Q-TOF series (Agilent Technologies, USA) or Waters Xevo series (Waters, USA). Reversed-phase highperformance liquid chromatography (RP-HPLC) semi-preparation was performed using a pentafluorophenyl column (Capcell-Pak PFP; 250 mm × 10 mm, 5 μm; Shiseido, Japan). A detailed description of the identification of compounds by means of a series of one-dimensional (1D) and two-dimensional (2D) nuclear magnetic resonance (NMR) analyses is provided in Appendix A.
《2.5. Methods for measuring antiviral activity》
2.5. Methods for measuring antiviral activity
2.5.1. Viruses, cells, and infection
Influenza strain A/Fort Monmouth/1/1947 (H1N1) was obtained from the America Type Culture Collection (ATCC, USA). Clinically isolated IAV strains A/tianjinjinnan/15/2009 (H1N1, oseltamivirresistant) and A/wuhan/359/1995 (H3N2) were kindly provided by Professor Yuelong Shu, at the National Institute for Viral Disease Control and Prevention, Chinese Centers for Disease Control and Prevention, China. Coronavirus HCoV-229E (VR-740) was purchased from the ATCC and HCoV-OC43 (VR-1558) was kindly provided by Dr. Xuesen Zhao, at Beijing Ditan Hospital, Capital Medical University, China. Human parainfluenza virus 3 (HPIV-3, strain C243) and respiratory syncytial virus (RSV, strain Long) were purchased from the ATCC.
Madin–Darby canine kidney (MDCK) and human hepatoma C3A cells were purchased from the ATCC. Human hepatoma Huh7.5, human cervical Hela, and Hep2 cells are kept in our laboratory. Antiviral assays against IAV were performed in the MDCK cells with a maintenance medium supplemented with 2 μg∙mL–1 tosyl phenylalanyl chloromethyl ketone (TPCK)-treated trypsin (Worthington, USA) and 0.08% bovine serum albumin (BSA; Yuan Heng Sheng Ma Biotech, China). IAV H3N2, if not specifically mentioned, was used throughout the anti-IAV study. Antiviral assays against other viruses were performed in Huh7.5 (HCoV-229E), C3A (HCoV-OC43), Hela (HPIV-3), and Hep2 (RSV) cells, and the maintenance medium was supplemented with 2% fetal bovine serum.
2.5.2. Cytopathic effect inhibition assay
The inhibitory activity of compounds against IAV, HCoV-229E, HPIV-3, and RSV was examined by means of a cytopathic effect (CPE) inhibition assay, as described previously [14]. In brief, cells seeded in plates at 37 °C overnight were infected with the virus at 100 times 50% tissue culture infective dose (TCID50) for 2 h (for HCoV-229E, the test compound was added simultaneously), followed by treatment with the test compounds for 48 or 72 h. The 50% inhibitory concentration (IC50) was determined by the Reed and Muench method [15]. The 50% toxicity concentration (TC50) of the test samples and positive compounds was also evaluated by the CPE assay. The selectivity index (SI) was calculated as the ratio of TC50/IC50.
2.5.3. qRT-PCR assay
The total RNA of the infected cells was extracted using the RNeasy Mini kit (Qiagen, Germany) according to the manufacturer’s instructions. The qRT-PCR was performed with TransScript II Green One-Step qRT-PCR SuperMix (TransGen Biotech, China) using the ABI 7500 Fast Real-Time PCR system (Applied Biosystems, USA) [16]. The primers used are listed in Table S2.
2.5.4. Western blot assay
In order to analyze the viral proteins, the cellular proteins were extracted using Mammalian Protein Extraction Reagent (M-PER; Thermo Fisher Scientific, USA) with a HaltTM protease inhibitor single-use cocktail. Immunoblotting for β-actin (antibody cat. No. 3700S; 1:5000; Cell Signaling Technology, USA) and IAV polymerase acidic (PA) protein (antibody cat. No. GTX118991; 1:1000; GeneTex, USA) was performed as described previously [16].
2.5.5. Time-of-addition assay
A time-of-addition experiment was performed as previously described [17], with some modifications. In brief, MDCK cells were inoculated with IAV for 2 h. The culture medium was replaced 2 h after IAV infection. The test compounds were added to the medium at indicated time points. The cells were harvested at 12 h post viral infection. Then, IAV PA protein was determined via western blot assay and the IAV titer was examined using CPE assay.
《2.6. Statistical analyses》
2.6. Statistical analyses
Data from the antiviral activity assay and qRT-PCR are presented as mean ± standard deviation (SD). Statistical analysis among the groups was conducted by the two-tailed Student’s t-test or one-way analysis of variance (ANOVA), followed by Dunnett’s correction, as applicable (GraphPad Prism software; GraphPad Software, USA); p values less than 0.05 were considered to be significant, and error bars denote the SD.
《3. Results》
3. Results
《3.1. Bioinformatic analysis of the genome sequence and activitydirected comparative transcriptomics analysis》
3.1. Bioinformatic analysis of the genome sequence and activitydirected comparative transcriptomics analysis
The fermentation broth and crude extracts from Streptomyces sp. 1647 were active against different strains of IAV, including H3N2, H1N1, and oseltamivir-resistant H1N1 (Table S3 in Appendix A). To demystify the structure of antiviral compounds, a multi-omics guided strategy, which differed from the conventional approach for the discovery of active compounds, was devised. This started with an analysis of the genome sequence of the producing strain (Methods part in Appendix A).
The genome of Streptomyces sp. 1647 is a linear chromosome of approximately 8.9 × 103 kb, with a GC content of 73.6%. The genome sequence was analyzed using antibiotics and Secondary Metabolite Analysis Shell Version 5.0.0 (antiSMASH 5.0.0) online tools↑ , which suggested that it contained 38 putative BGCs for the production of secondary metabolites.
↑ http://antismash.secondary metabolites.org
Comparative transcriptome analysis (Methods part in Appendix A) was conducted to preliminarily locate the BGC responsible for the biosynthesis of the antiviral active compounds in Streptomyces sp. 1647. Through the screening of 14 types of fermentation media, A3 and B7 were selected as highly active and inactive fermentation media, respectively. The mycelia and fermentation supernatants from these two media at the early stages of fermentation (24, 48, and 72 h) were collected to perform transcriptome sequencing (RNA-seq) (Fig. 1(a)). In conjunction with the information of 38 secondary metabolite BGCs predicted by antiSMASH, three BGCs—namely, clusters 27, 28, and 36—showed significantly higher transcription levels under the A3 fermentation condition (Figs. 1(b)–(d)). The transcriptomic results were further verified by qRT-PCR (Figs. S1(a)–(c) in Appendix A).
《Fig. 1》
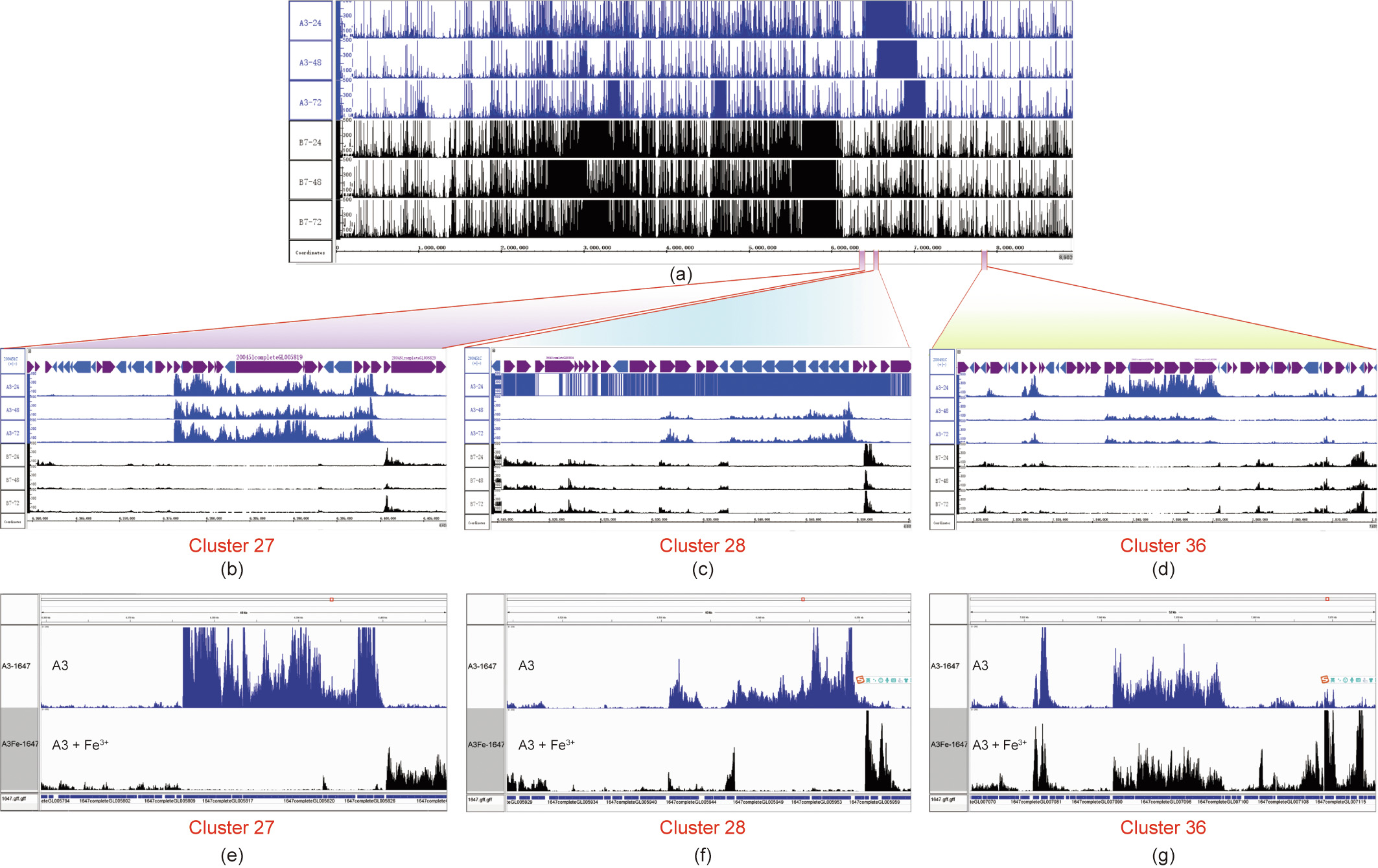
Fig. 1. Transcriptome alignment of Streptomyces sp. 1647 under different fermentation conditions. The mycelia of the wild-type (WT) strain from two media (A3 and B7) at the early stage of fermentation (24, 48, and 72 h, respectively) were collected for the extraction of total RNA; (a)–(d) these six samples were then subjected to RNA-seq. (e)–(g) The mycelia of Streptomyces sp. 1647 cultured by A3 and A3 + Fe3+ (FeCl3 was added to a final concentration of 0.05%) media were collected at 48 h for the extraction of total RNA and RNA-seq analyses. (a) Genome-wide transcriptome alignment; (b) transcriptome alignment at cluster 27; (c) transcriptome alignment at cluster 28; (d) transcriptome alignment at cluster 36; (e) gene transcription levels of cluster 27 in the presence of Fe3+; (f) gene transcription levels of cluster 28 in the presence of Fe3+; (g) gene transcription levels of cluster 36 in the presence of Fe3+. The alignments from A3 medium are shown as blue peaks, and those from B7 and A3 + Fe3+ media are displayed as black peaks.
Based on bioinformatic analysis, both clusters 27 and 28 were found to be BGCs for the production of siderophores, as the presence of iron box sequences in the cluster, which were closely regulated by a class of repressors in Streptomyces, sensed the presence of Fe3+ [18,19]. The transcriptome analysis demonstrated that clusters 27 and 28 were shut down after adding a sufficient amount of Fe3+ (0.05% final concentration) to the A3 fermentation medium, while cluster 36 was still highly expressed (Figs. 1(e)– (g)). This was verified by qRT-PCR (Figs. S1(d)–(f) in Appendix A). The activity assay showed that Streptomyces sp. 1647 fermentation broth in the presence of Fe3+ still exhibited a similar level of antiIAV activity (Table S3). Thus, we speculated that cluster 36 was the BGC responsible for the biosynthesis of antiviral active compounds in Streptomyces sp. 1647.
《3.2. Confirmation of the BGC for the biosynthesis of antiviral active compounds》
3.2. Confirmation of the BGC for the biosynthesis of antiviral active compounds
As predicted by antiSMASH 5.0.0, cluster 36 contains 50 open reading frames (ORFs) with some non-ribosomal peptide synthetase (NRPS) genes (Fig. 2(a)). To preliminarily evaluate the role of cluster 36 in the biosynthesis of the antiviral metabolites in Streptomyces sp. 1647, we performed overexpression of multiple regulatory genes situated in this cluster. The five regulatory genes—namely, 7081 (TetR family), 7082 (streptomyces antibiotic regulatory protein (SARP) family), 7083 (ArsR family), 7089 (LysR family), and 7102 (TetR family)—near the NRPS genes in this cluster were selected, and the results revealed that only the overexpression of gene 7102 caused an increase in the anti-IAV activity (Fig. 2(b) and Figs. S2(a)–(d) in Appendix A). The gene expression changes of cluster 36 between the wild-type (WT) strain and gene 7102 overexpression strain shown by the RNA-seq data suggested that overexpression of gene 7102 did indeed cause a significant up-regulation of the genes 7092–7102 in the NRPS core region of cluster 36 (Fig. 2(c)). This finding was confirmed by qRT-PCR (Fig. S2(e) in Appendix A). The proposed function of the genes 7092–7102 at the core NRPS region regulated by gene 7102 is given in Table S4 in Appendix A.
《Fig. 2》

Fig. 2. Analysis of the overexpression and knockout mutants of Streptomyces sp. 1647. (a) Schematic representation for five regulatory genes (marked by green arrows) and NRPS gene 7098 (marked by a red arrow) in cluster 36. (b) Inhibitory activities of the fermentation supernatant samples collected from the WT strain and gene 7102 overexpression strain (7102) cultured in A1, A2, and A3 + Fe3+ media. FeCl3 was added to the A3 medium to a final concentration of 0.05% to obtain the A3 + Fe3+ medium. The 1/10 dilution of fermentation broth was further three-fold diluted seven times, and then virus-induced CPE was recorded to give IC50. (c) Transcriptome alignment results of cluster 36 between the WT strain (black) and gene 7102 overexpression strain (blue, 1647/pL-7102) cultured in A1 medium. (d) The qRT-PCR analysis of gene expression levels in the core NRPS region of cluster 36 in the WT strain, 7098 gene knockout mutant (7098-KO), and genetically complementary strain (7098-KOC). (e) Inhibitory activities of the fermentation samples collected from the strains WT, 7098-KO, and 7098-KOC cultured by A3 + Fe3+ medium. (f) LC–MS analysis of the fermentation crude extracts obtained from the above strains. The purple-shaded region represents the products of cluster 36. Con: treated with equal amount of vehicle. The experiments were performed in triplicate, and each value represents the mean ± SD. Student’s t-test: **p < 0.01, ***p < 0.001 vs the WT strain.
After this result, knockout of the NRPS gene 7098 in the core region of cluster 36 by an in-frame deletion was conducted. The genetically complementary strain 7098-KOC was obtained by reintroducing gene 7098 into mutant 7098-KO to avoid any polar effect of the deletion. The gene expression level of the core region of cluster 36 was detected by qRT-PCR (Fig. 2(d)), and the antiviral activity test (Fig. 2(e)) showed that inactivation of gene 7098 led to a loss of anti-influenza virus activity, while complementation of this gene could restore the antiviral activity. LC–MS analysis (Fig. 2(f)) of the fermentation extracts from these strains indicated that a series of compounds with retention times at 14–23 min and 31– 42 min disappeared in the knockout mutant 7098-KO, and then reappeared in the complementary strain 7098-KOC.
These results demonstrated that the core NRPS region of cluster 36 was responsible for the biosynthesis of the active substances against IAV produced by Streptomyces sp. 1647. During this study, the cluster responsible for deimino-antipain was reported by Maxson et al. [20] using a chemical reactivity-based screening. This is highly homologous to the core region of cluster 36. These data indicate that the active agents against IAV might be oligopeptides with different post-modifications as compared with deiminoantipain.
《3.3. Activity-based comparative metabolomics analysis》
3.3. Activity-based comparative metabolomics analysis
To efficiently identify secondary metabolites with antiviral activity in Streptomyces sp. 1647, a molecular networking approach was applied. This was based on the global fragmentation pattern profile obtained by electrospray HR-ESI-MS/MS after the HPLC separation of metabolites in the WT strain, 7098 gene blocked mutants (7098-KO), and genetically complementary strains (7098-KOC). The fermentation crude extracts of these strains were divided into three groups: G1, G2, and G3.
The MS/MS data of G1, G2, and G3 were submitted to the GNPS molecular networking database, an MS system-based online platform [21,22] (Fig. 3(a)). A total of 22 850 mass spectra were clustered to form 1076 nodes in this molecular networking, and each node represented the MS/MS data of structurally similar compounds. A special molecular cluster was formed by compounds from groups G1 and G3 together, but not from the inactive group G2 (Fig. 3(b)). This suggested that the molecules in these nodes were most likely to belong to the antiviral active compounds. The characteristics of compounds in this molecular cluster, including the distribution information, molecular weight, ultraviolet (UV) absorption, and MS/MS fragmentary ions data, were applied to facilitate the isolation and structural elucidation of the target antiviral compounds.
《Fig. 3》
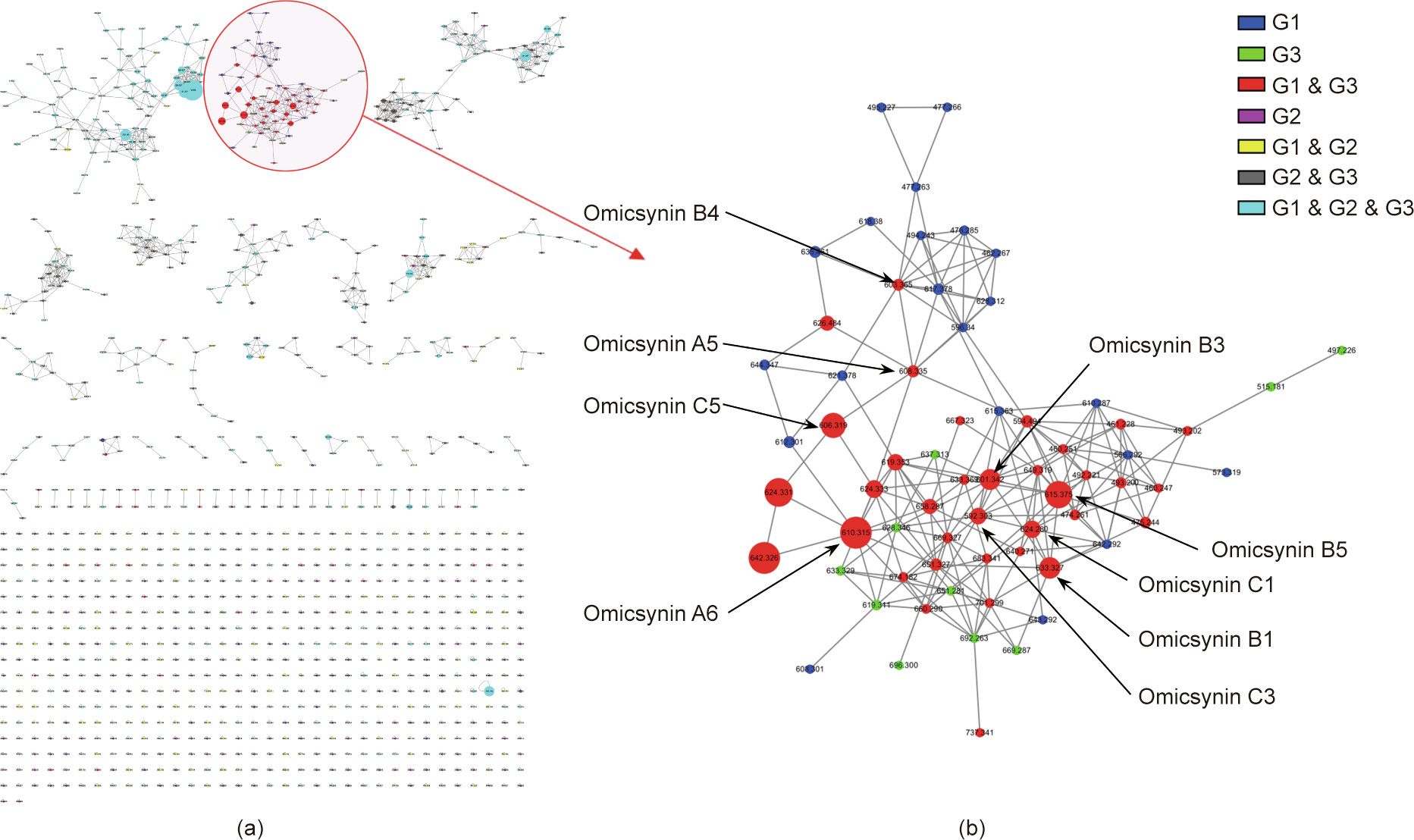
Fig. 3. GNPS molecular networking based on the MS/MS fragmentation data of fermentation crude extracts. (a) A cluster view of the molecular networking of the MS/MS fragmentation data of fermentation crude extracts from the WT strain (group G1), gene 7098 knockout mutants (group G2), and genetically complementary strains (group G3) was constructed by GNPS online workflow. A cluster of components that only occurred in the WT strain and the genetically complementary strain (groups G1 and G3) with antiviral activity are displayed with a red circle and light shade. (b) Node view for the special cluster for antiviral compounds represented by the molecular weight of the parent ions in negative mode.
《3.4. Isolation and structural determination of the antiviral metabolites》
3.4. Isolation and structural determination of the antiviral metabolites
To obtain sufficient amounts of active compounds for full structure elucidation, a large-scale fermentation of the WT strain of Streptomyces sp. 1647 was conducted, with the gene 7098 knockout mutant as a negative control. Guided by the analysis data of molecular networking and antiviral activity, 18 pseudotetrapeptides with an internal ureido linkage were identified and named as omicsynins. They were omicsynins A1–A6, omicsynins B1–B6, and omicsynins C1–C6, with their C-terminal structures being phenylalaninol (Pheol), arginal (Argal), and phenylalaninal (Pheal), respectively. Among them, A1, A2, A6, B1–B3, B5, B6, C1, C2, and C6 are new compounds (Fig. 4(a)).
《Fig. 4》

Fig. 4. Structural information of omicsynins. (a) The structures of omicsynins A1–A6, B1–B6, and C1–C6. (b) The structures, key 1H–1H correlation spectroscopy (COSY), heteronuclear multiple bond correlation (HMBC) spectroscopy, and nuclear Overhauser effect spectroscopy (NOESY) correlations of omicsynins A1–A4. Cap: capreomycidine; Met: methionine; THPI: 4-substituted tetrahydropyrimidin-2(1H)-imine; MTE: methylthioethyl; Bn: benzyl; Arg: arginine; GP: guanidinopropyl; Val: valine; i-Pr: isopropyl; i-Bu: isobutyl; s-Bu: sec-butyl; Leu: leucine; Ile: isoleucine; novel: novel compound; MAPI: microbial alkaline protease inhibitor.
Omicsynin A1 was isolated as a white amorphous powder, and its molecular formula was determined to be C30H41N7O6S by means of HR-ESI-MS ([M + H]+ m/z 628.2903, Calcd. 628.2912). The NMR data analysis revealed its structure of phenylalanine (Phe)–CO– capreomycidine (Cap)–methionine (Met)–Pheol (Fig. 4(b) and Fig. S3 and Table S5 in Appendix A), with a hydroxide at the Cterminal Phe residue. The HR-ESI-MS data of omicsynin A2 indicated a molecular formula of C30H43N7O6S, which has two hydrogens more than A1. According to the NMR data, omicsynin A2 was determined to be Phe–CO–arginine (Arg)–Met–Pheol (Fig. 4(b) and Fig. S4 and Table S5 in Appendix A). The absolute configuration of omicsynins A1 and A2 was determined using acid hydrolysis and advanced Marfey’s method [23,24] (Table S6 in Appendix A). Omicsynins A3 and A4 were identified as known compounds by means of NMR data analysis (Fig. 4(b)) and by comparison with the literature [25,26]. Comprehensive analysis of the HR-ESI-MS/MS data (Fig. S5 in Appendix A) of omicsynins A3–A6 demonstrated that the Val residue in A3 and A4 was replaced by a leucine (Leu) or isoleucine (Ile) unit in the omicsynins A5 and A6.
The C-terminal forms of omicsynins B1–B6 were identified as aldehydes of Arg residue (Argal) based on the 1H-NMR data and HR-ESI-MS/MS analysis, along with the HPLC-based co-injection analysis of omicsynin B4 and antipain (EFEBIO, Shanghai, China), a known peptide-aldehyde protease inhibitor (Fig. S5). Antipain exists as a dynamic equilibrium mixture of two hydrate forms, four cyclic carbinolamine forms, and two unhydrated aldehyde forms in aqueous solutions [27]. The instability of Argal led to the chromatographic difficulties previously encountered, as well as the unavailability of high-quality 2D NMR spectra. Omicsynins C1–C6 varied by –2 Da from omicsynins A1–A6, respectively, and the C-terminal of the omicsynin Cs was speculated to be an aldehyde based on the HR-ESI-MS/MS data (Fig. S5) and on a comparative analysis with the commercial compound chymostatin.
《3.5. Antiviral activity of the omicsynins》
3.5. Antiviral activity of the omicsynins
Omicsynins A1–A4 and B1–B4, together with the commercially available antipain and chymostatin, were tested for their activity against IAV and coronavirus HCoV-229E by CPE assay (Table 1). Omicsynins B1–B4 exhibited significant antiviral activity against IAV, with IC50 values in the low micromolar concentration range (0.89–3.34 μmol·L–1), which were more potent than those of oseltamivir phosphate (IC50 4.24 μmol·L–1) and RBV (IC50 13.16 μmol·L–1). Similarly, omicsynins B1–B4 possessed significant anti-HCoV-229E effects with IC50 values at about 1 μmol·L–1, which were about 20 times more potent than that of the positive reference RBV (IC50 25.14 μmol·L–1). Omicsynins A1–A3 exhibited moderate anti-IAV (IC50 80–320 μmol·L–1) and anti-coronavirus activities (IC50 40–180 μmol·L–1); chymostatin also showed moderate inhibitory activity against coronavirus HCoV-229E (IC50 23.74 μmol·L–1), which was more than ten times higher than its activity against IAV (IC50 329.33 μmol·L–1). These results suggested that the C-terminal reduction forms (i.e., aldehyde or alcohol hydroxyl) and the C-terminal amino acid (i.e., Arg or Phe) were of significant importance to the antiviral activities and provided early information that could be useful for future chemical optimization.
《Table 1》
Table 1 Anti-IAV and HCoV-229E activity of the active compounds from Streptomyces sp. 1647.
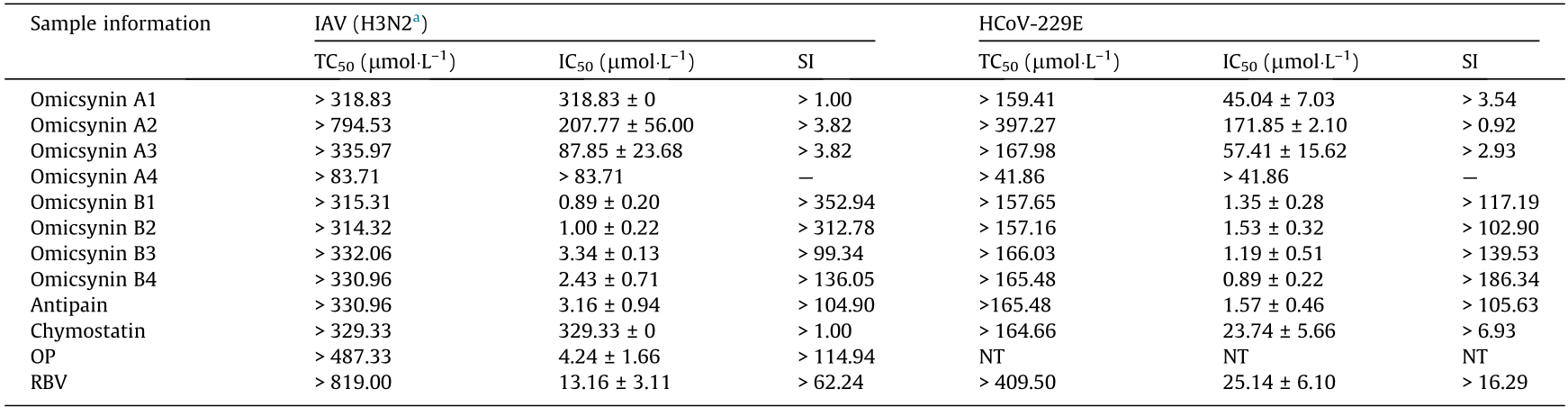
Antipain was equivalent to the compound omicsynin B4. Chymostatin contained three compounds, chymostatin A, B, and C, which were equivalent to the compounds omicsynin C3 and C5. Oseltamivir phosphate (OP; Roche, China) and ribavirin (RBV; Sigma-Aldrich, USA) were used as positive control compounds. NT: not tested. n = 3, the data represents the mean ± SD.
a The GenBank accession numbers of the H3N2 virus are CY112821.1–CY112828.1.
To further investigate the antiviral spectrum of the omicsynins, the antiviral activity toward other respiratory RNA viruses, including HCoV-OC43, RSV, and HPIV, was determined. CPE inhibition analysis revealed that the omicsynins had no antiviral activity against RSV and HPIV (data not shown). The inhibitory activity of omicsynin to HCoV-OC43 was detected by the qRT-PCR analysis of HCoV-OC43 N protein messenger RNA (mRNA) in C3A cells, since HCoV-OC43 infected C3A cells without inducing CPE. The results showed that omicsynin B4 exhibited inhibitory activity against coronavirus HCoV-OC43, with an IC50 of 28.67 μmol·L–1 (Fig. 5(a)).
The antiviral activity of omicsynin B4, the fermentation extract from Streptomyces sp. 1647 WT strain (WT-50E) and the knockout mutant of gene 7098 (KO-50E) were further examined at the levels of IAV M2 mRNA and PA protein, with RBV as a positive control. As shown in Figs. 5(b) and (c), the omicsynin B4 and WT-50E treatments dose-dependently reduced the levels of IAV M2 mRNA and PA protein in MDCK cells. However, KO-50E did not affect the replication of IAV.
To investigate which step of IAV replication is targeted by omicsynins, a time-of-addition experiment [17] was performed, as shown in Figs. 5(d) and (e). It was found that omicsynin B4 and WT-50E treatment could inhibit the IAV titers after viral infection (2–12 h) rather than before (–2–0 h) and during (0–2 h) the infection (Fig. 5(d)). Furthermore, it was found that omicsynin B4 and WT-50E could inhibit the IAV replication when added at 0, 1, 2, 4, or 6 h post-infection, but not at 8 and 10 h post-infection (Fig. 5(e)). Meanwhile, KO-50E had no obvious antiviral effect on all stages of viral replication, as expected. Taken together, the results indicate that omicsynin B4 and WT-50E inhibit the early stage of viral replication through post-entry events.
《Fig. 5》
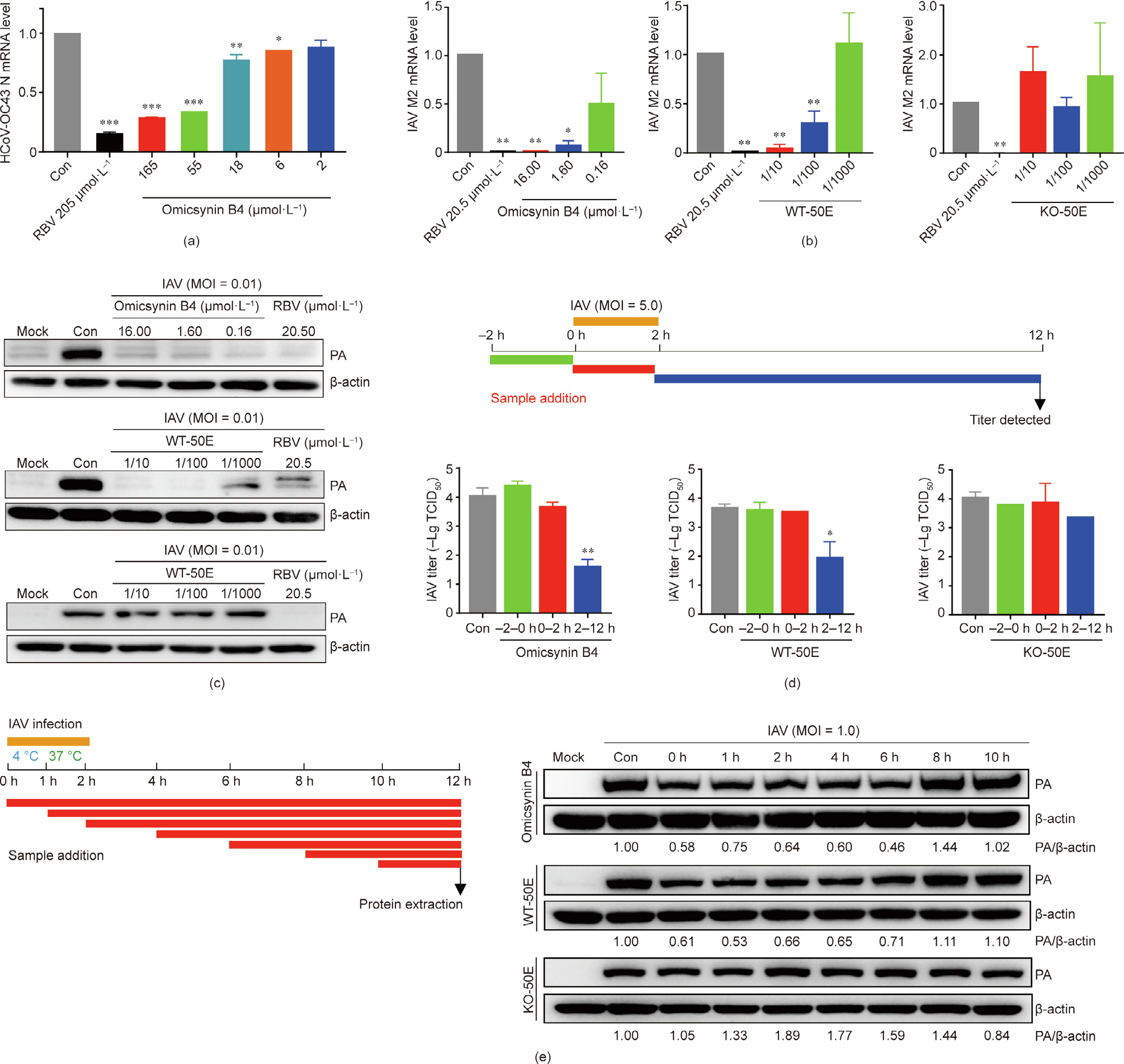
Fig. 5. Inhibition assay of omicsynins against HCoV-OC43 and IAV H3N2. (a) The mRNA expression levels of the N protein of HCoV-OC43 in C3A cells, determined by qRT-PCR assays. (b, c) Effects of omicsynin B4, WT-50E, and KO-50E on IAV M2 mRNA and PA protein level in MDCK cells. MDCK cells were infected with IAV H3N2 (multiplicity of infection (MOI) = 0.01) and test samples were added at 2 h post-infection. After 24 h post-infection, the cells were harvested and (b) IAV M2 mRNA was determined by qRTPCR assay and (c) viral PA protein was assayed by Western blot assay. (d, e) Time-of-addition assay of omicsynin B4, WT-50E, and KO-50E against IAV. MDCK cells were infected with IAV H3N2 and tested samples were added at the indicated time, as presented in the schematic experimental design. After 12 h post-infection, the cells were harvested and the viral titer was determined by (d) CPE assay and (e) Western blot assay of the viral PA protein. WT-50E and KO-50E are the fermentation extracts of Streptomyces sp. 1647 WT strain and the knockout mutant of gene 7098. Con: treated with equal amount of vehicle. Each value represents the mean ± SD. One-way ANOVA: *p < 0.05, **p < 0.01, ***p < 0.001 vs Con.
《4. Discussion》
4. Discussion
Microorganisms are constantly exposed to viruses (for prokaryotes, bacteriophages) in nature. Therefore, microorganisms may produce metabolites that provide a microenvironment that is resistant to viral infection. Among the more than 20 000 fermentation samples screened in our institute in past decades, Streptomyces sp. 1647 was discovered to be highly active and repeatable. It produces secondary metabolites with good antiviral activity to IAV, including an oseltamivir-resistant strain. Intriguingly, the active components from the fermentation broth were reported to be active against SARS-CoV [28]. However, the identification of the antivirus compounds produced by Streptomyces sp. 1647 has long puzzled researchers. Here, we used an integrative ‘‘omics” strategy to identify a gene cluster responsible for the biosynthesis of the antiviral secondary metabolites produced by the Streptomyces sp. 1647 and ultimately determined the corresponding active antiviral compounds, 11 of which are novel.
In the traditional microbial natural product discovery process, the isolation and identification of active natural compounds is laborious and time-consuming. Sometimes, the active compounds may be lost or inactivated after a series of bioassay-guided isolations and separations, although the initial microbial fermentation extracts may show good bioactivity. Due to the lack of clear NMR data for hydrogen and carbon signals, the entire molecular structure may not be elucidated or may be erroneously determined. Thus, a novel strategy was developed by integrating the cuttingedge techniques of genomics, bioactivity-guided transcriptomics, and metabolomics analysis. This approach contributed to the identification of a series of novel antiviral pseudo-tetrapeptide natural products named omicsynins, produced by Streptomyces sp. 1647. The highly active omicsynin Bs compounds with C-terminal Argal might exist primarily in their hemiaminal form with minor amounts of free aldehyde and hydrate in weakly acidic aqueous solution [20]. This likely contributed to the difficulties encountered in the activity-guided separation procedures and HPLC analysis.
In the post-genomics era, multi-omics analyses can be effectively used to facilitate the discovery and identification of bioactive compounds from microbial natural products. Efficient bioinformatic analysis based on genome sequences may yield primary structural information on the secondary metabolites encoded by the target strain. Furthermore, the applications of bioactivitybased comparative transcriptomics may determine the potential BGCs responsible for the biosynthesis of active compounds. The target BGC may be further determined by genetic manipulation of the regulatory or biosynthetic genes in the cluster. With the structural information predicted from the target BGC, the bioactivity-oriented molecular networking of metabolomics data may highlight the putative bioactive candidates produced by the strain. Thus, rationalized isolation procedures may be adopted according to the untapped characteristics of target molecules, which may accelerate the bioactive compound identification process [29,30], especially for intractable compounds, using traditional separation procedures.
This novel strategy has another inherent advantage derived from the omics data—that is, the simultaneous discovery of the BGC and its active secondary metabolites. The BGC responsible for omicsynin production is located in the core NRPS region of cluster 36 encoding five NRPS genes. It is highly homologous with the BGC of deimino-antipain (Fig. 6(a)). Deimino-antipain belongs to a family of peptide-aldehyde protease inhibitors characterized by the presence of C-terminal aldehydes [31–33], including antipain, chymostatin, β-microbial alkaline protease inhibitor (β-MAPI), elastatinal A, and leupeptin (Fig. 6(b)). In Streptomyces sp. 1647, more than 18 omicsynins are identified by metabolomics data, encompassing at least 11 novel compounds, as well as several known analogues, such as antipain, chymostatin, and β-MAPI, which have been reported to possess multiple antiviral activities. For example, antipain has been shown to have inhibitory activities against HCoV-229E [34] and poliovirus 2A [35], leupeptin against influenza virus [36] and HCoV-229E [34], and β-MAPI against HIV-1 protease [26,36]. The novel peptide-aldehyde compounds omicsynin B1, B2, and B3 exhibited significant inhibitory activities against IAV and HCoV-229E, acting in a broad-spectrum fashion. Omicsynin B4 and multiple omicsynin components exist in the 50E part of the WT strain and could inhibit IAV replication when added from 0 to 6 h after infection, which suggests that they exert anti-IAV effects through interference with the early stage of postentry events. Similar activity of omicsynins on the oseltamivirsensitive and resistant IAV strain suggests that their antiviral mechanism does not involve targeting the neuraminidase, which facilitates the release of progeny viruses from infected cells. The exact role of the omicsynins involved in the anti-IAV mechanism warrants further detailed investigation.
《Fig. 6》
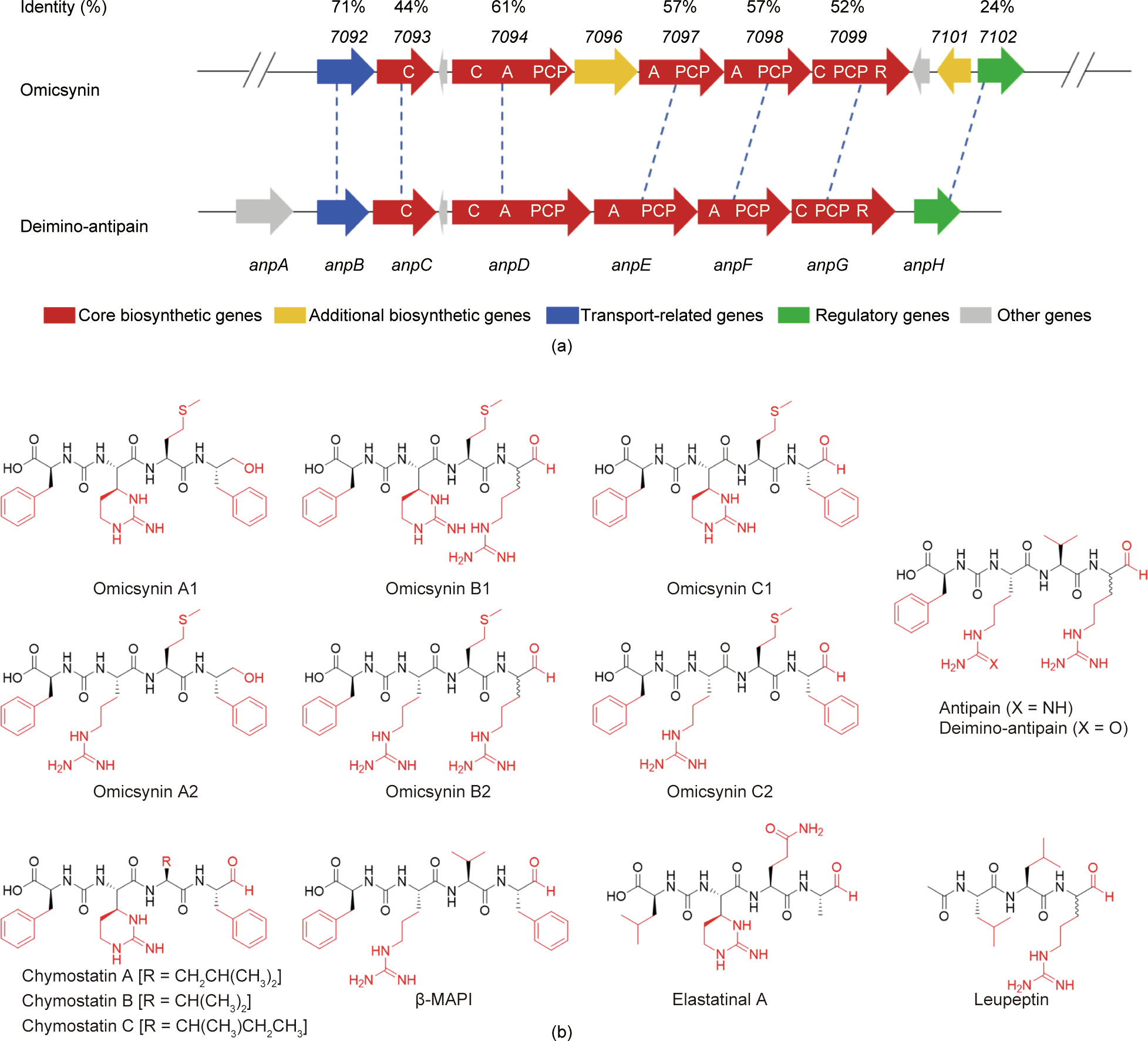
Fig. 6. Genetic organizations and chemical structures of omicsynins and representative peptide-aldehyde protease inhibitors. (a) Schematic representation of the omicsynin gene cluster and the deimino-antipain gene cluster, and their amino acid sequence identities. (b) Chemical structures of omicsynins and representative peptide-aldehyde protease inhibitors including deimino-antipain, antipain, chymostatin, β-MAPI, elastatinal A, and leupeptin.
This is the first report to identify an antiviral gene cluster simultaneously with the discovery of corresponding chemicals in microorganisms. However, the biosynthetic mechanism of these compounds has not yet been elucidated. There are a number of unsolved mysteries in the biosynthesis process of omicsynins, such as the unusual lack of the fourth NRPS A domain (only three A domains are present in the cluster for tetrapeptide production) and the functions of two unique oxidoreductases. The presence of congeners of this pseudo-peptide family suggests the presence of various genetic elements for the biosynthesis of novel antiviral chemicals. Thus, determination of the detailed biochemical machinery for omicsynin biosynthesis and the genome mining of new biosynthetic modules pertaining to omicsynin and its structural analogues will facilitate the discovery of novel derivatives through the means of combinatorial biosynthesis and synthetic biology. This may lead to new drug candidates or therapies for fighting the respiratory infections caused by influenza virus and/ or coronavirus.
《5. Conclusions》
5. Conclusions
We used an integrated multi-omics strategy to discover more than ten novel antiviral pseudo-tetrapeptides—namely, omicsynins produced by Streptomyces sp. 1647. The omicsynins, especially the omicsynin Bs, exhibit significant antiviral activities against IAVs and coronaviruses. The simultaneous identification of the BGC provides insights into the genetic mechanisms involved in the biosynthesis of omicsynins; it also provides the potential for discovering new antiviral compounds to cope with outbreaks of IAV and coronavirus-related diseases.
《Acknowledgments》
Acknowledgments
This work was supported by the National Natural Science Foundation of China (81630089, 81703398, 81872780, and 81803410), the Beijing Natural Science Foundation, China (7214286), the Drug Innovation Major Project of China (2018ZX09711001-006-011, 2018ZX09735001-002, and 2018ZX09711001-007), and the CAMS Innovation Fund for Medical Sciences (2018-I2M-3-005 and 2020- I2M-2-010).
《Authors’ contribution》
Authors’ contribution
Shuyi Si, Bin Hong, Jian-Dong Jiang, and Yuhuan Li contributed to the study design. Hongmin Sun, Xingxing Li, Minghua Chen, Ming Zhong, Yihua Li, Kun Wang, Yu Du, Xin Zhen, Rongmei Gao, Yexiang Wu, and Yuanyuan Shi contributed to the experiment implementation and the data collection and analysis. Shuyi Si, Bin Hong, Jian-Dong Jiang, Yuhuan Li, Hongmin Sun, Xingxing Li, Minghua Chen Liyan Yu, and Yongsheng Che contributed to the manuscript preparation. All authors have read and approved this final manuscript.
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Hongmin Sun, Xingxing Li, Minghua Chen, Ming Zhong, Yihua Li, Kun Wang, Yu Du, Xin Zhen, Rongmei Gao, Yexiang Wu, Yuanyuan Shi, Liyan Yu, Yongsheng Che, Yuhuan Li, Jian-Dong Jiang, Bin Hong, and Shuyi Si declare that they have no conflict of interest or financial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2021.05.010.














 京公网安备 11010502051620号
京公网安备 11010502051620号




