Two million years ago, in what is now the icy barren tip of northern Greenland, mastodons, reindeer, rodents, and geese roamed through forests of poplar, spruce, and birch. The existence of that surprisingly lush ecosystem is one conclusion of research reported in a December 2022 Nature paper in which an international team of scientists sequenced 2-million-year-old plant and animal deoxyribonucleic acid (DNA) isolated from frozen sediment samples [1]. Their work not only furthered research on evolution and climate change, but shattered the previous record for ancient DNA reconstruction, set in 2021 when another group reported on their sequencing of 1.2-million-year-old mammoth DNA [2].
‘‘We are now at the point in ancient DNA reconstruction where we are not limited by technology, but by the practical limit of how long DNA can survive under perfect, frozen permafrost conditions,” said Love Dalén, professor in evolutionary genetics at Stockholm University in Sweden and leader of the study on mammoths that was the first to recover DNA beyond the million-year barrier. ‘‘At least in the Northern Hemisphere, there is no permafrost older than about 2.6 million years.”
Researchers are approaching this ancient limit thanks to an improved ability to extract small, degraded pieces of DNA from samples, sequence those fragments, and computationally assemble those data into larger genetic chunks. The price of DNA sequencing has also dramatically decreased over the last decade, with the cost of whole human genome sequencing now approaching just 100 USD [3]. This makes it more affordable for paleogeneticists to sequence more of the ancient DNA they collect. ‘‘In the past, when it was more expensive, you really had to only focus on your highquality samples and longer segments of DNA,” Dalén said. ‘‘Now, we can generally afford to dig more deeply into samples and sequence everything.”
Researchers have also developed new buffers and extraction methods to remove DNA more gently and effectively from sediment or decomposed tissue. The extraction methods used to reconstruct the DNA from ancient Greenland, for instance, were newly optimized based on a series of experiments studying how DNA adheres to clay, silicate, and quartz particles found in permafrost [1].
At the same time, the advent of new DNA sequencing technologies and accompanying bioinformatics tools are allowing paleogeneticists to sequence and assemble smaller fragments of DNA than ever before. Not long ago, researchers working with short DNA fragments struggled to be sure which small bits came from the species being analyzed, and which were contaminants, Dalén said. His team, though, showed it was possible to use DNA fragments as short as 35 base pairs long when studying the 1.2-million-yearold Siberian mammoths (Fig. 1) [2]. That is in large part, he said, due to new alignment and mapping techniques—the final step where millions or billions of short DNA sequences are pieced together like a puzzle to spell out entire genes and genomes. If this is not done correctly, the resulting DNA sequence can be jumbled or include small bits of contaminating bacterial DNA in addition to the plant, animal, or human DNA of interest.
《Fig. 1》

Fig. 1. Recent advances in genomic science are letting researchers better isolate, sequence, and analyze ancient DNA samples, providing new insights into the evolution of ancient humans, plants, and animals like these woolly mammoths, shown in an artist’s illustration. Credit: Beth Zaiken, with permission.
In the work analyzing the more-than-1-million-year-old Krestova mammoth, Dalén’s group employed an iterative mapping approach, using multiple rounds of mapping to determine how the DNA sequences fit together. Their final genome was detailed enough that they could detect, among other things, the genes that gave the mammoths their woolly coats and let them adapt to the cold [4]. These results have provided insights into how and when mammoths began populating cold regions of the globe.
A plethora of other paleogenomic research is also providing new insights about the earliest humans, with genomic sequencing of bones from tens of thousands of years ago providing new pieces of the story about how, when, and where modern human populations emerged. ‘‘As the technology to analyze ancient DNA keeps improving, we have a better and better understanding of what people experienced in the past,” said He Yu, an assistant professor of life sciences at Peking University in Beijing, China. ‘‘What happened during these time periods is important not only for understanding the history of humankind, but for answering broader questions about population genetics, evolution, and climate change.”
Yu was part of an international team of scientists that in March 2023 reported a genomic analysis of the remains of 356 ancient hunter-gatherers from 35 000 to 5000 years ago (Fig. 2) [5]. Based on their results, the researchers concluded that, during the peak of the last ice age, ancient Europeans survived in the relatively warm areas of southwestern Europe. After the last glacial maximum, 25 000 to 19 000 years ago, those populations then expanded throughout the rest of Europe. ‘‘For a long time, people wondered how these lines of ancestry were preserved; they seemed to disappear for over 10 000 years and reappear again,” Yu said. ‘‘We found this ancestry did not disappear at all; they were always preserved in western Europe and previous studies did not capture that.”
《Fig. 2》
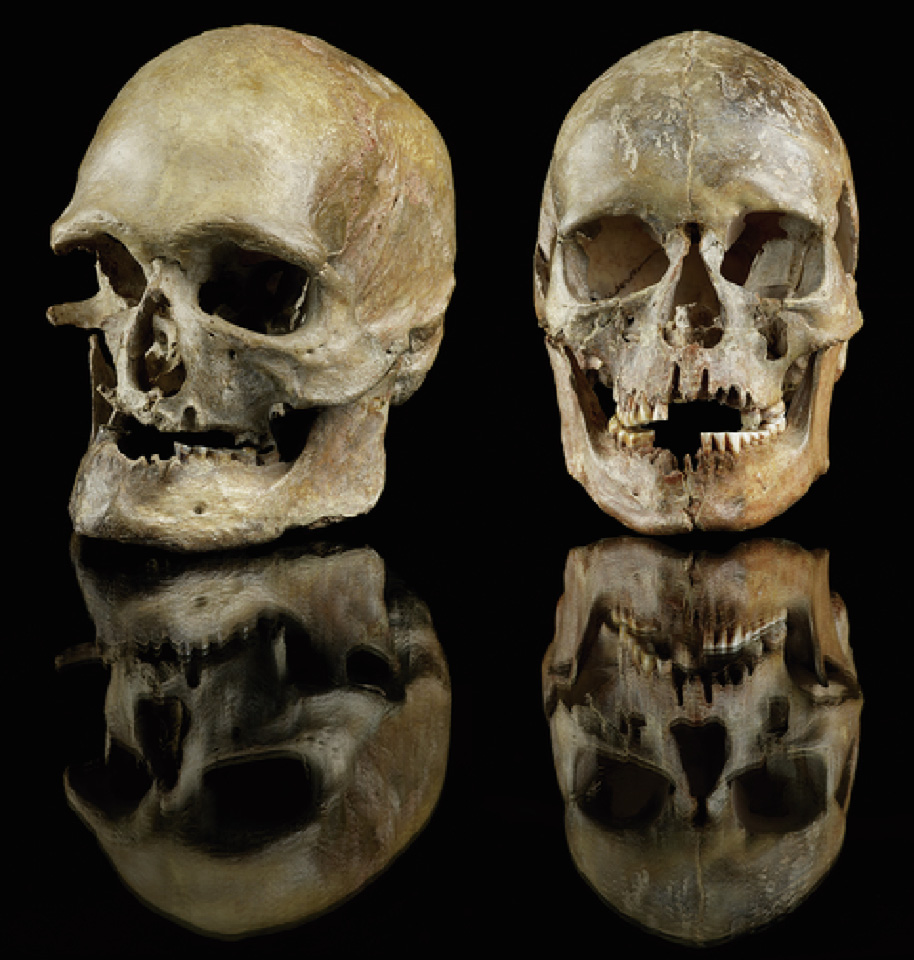
Fig. 2. Buried in Germany about 14 000 years ago, these skulls were among the remains of 356 prehistoric hunter-gatherers from different cultures whose ancient DNA was sampled and analyzed to study how and when modern humans spread across Eurasia. Credit: Jürgen Vogel, LVR-LandesMuseum Bonn, with permission.
Yu added that a relatively new method of preparing ancient DNA for sequencing allowed her and her collaborators to capture more information than in the past. In places where doublestranded DNA was damaged and had ragged, single-stranded ends—a common finding in old, degraded DNA samples— researchers previously cut off and discarded these overhangs. Yu and her colleagues used a method that instead retained them by separating all the DNA into single strands [6]. ‘‘This approach, just described in the last few years, lets us sequence more of our short ancient DNA fragments,” she said.
Similar studies on ancient humans are now becoming abundant. In May 2022, a research team led by Laurent Excoffier, professor of population genetics at the University of Bern in Switzerland, reported a genomic analysis of the remains of 25 humans from several archaeological sites across Southwest Asia and Europe; the results allowed them to posit a time and place for the dawn of farming [7]. In March 2023, scientists at the Chinese Academy of Sciences in Beijing reported new genomic data from the ancient remains of 89 humans found at 29 archaeological sites spanning the Tibetan Plateau; the data added to the historical understanding of how and when populations across the plateau crossed paths with each other [8]. The same month, other researchers provided a new timeline of when Asian and African human lineages first mixed, based on their genomic analysis of ancient DNA from remains of members of the Swahili civilization excavated from cemeteries in Kenya [9].
The same technological improvements allowing these views into the distant past are also providing new data on more recent human history. In June 2022, for instance, scientists in Germany and Scotland published the results of their research sequencing DNA from the teeth of people buried in the 14th century to uncover new clues about where the bubonic plague pandemic known as the Black Death originated [10]. The team pieced together fragments of bacterial DNA still present in the teeth of people who died in Kyrgyzstan a few years before the Black Death swept through Europe, killing as many as 200 million people. This DNA confirmed which bacterial strain caused the plague, suggested that it jumped from marmots to humans, and showed how the pandemic likely began in the mountainous Kyrgystan region in Central Asia before spreading throughout Eurasia [10,11].
Shortly after that paper was published, in October 2022, a separate research group published their genomic analysis of DNA from the remains of people buried in London mass graves during the Black Death (Fig. 3). Their results added to the story of how the plague shaped human history, pinpointing a gene that provided protection against the plague; the gene became more common in both London and Denmark after the Black Death [12]. ‘‘This was a very direct way to evaluate the impact that a single pathogen had on human evolution,” said Luis Barreiro, professor of genetic medicine at the University of Chicago (Chicago, IL, USA) and cosenior author on the paper. He added that the study could not have been performed without recent advances in sequencing technology [13].
《Fig. 3》

Fig. 3. To learn more about how the Black Death plague pandemic in the mid-1300s shaped the human genome, researchers isolated and analyzed DNA from bodies buried in the East Smithfield plague pits in London, which were used for mass burials in the years 1348 and 1349. Credit: Museum of London Archaeology (public domain).
Eske Willerslev, professor of geogenetics at the University of Copenhagen, the Netherlands, and leader of the research team sequencing DNA from permafrost in Greenland, has firsthand experience with how transformational the advances in technology have been for genomics-based research. He has said that he had been trying to sequence the same soil samples since 2006 [14].
‘‘Every time we made improvements in our DNA extraction methods or our sequencing abilities, we went back to the samples and tried again,” Willerslev recalled [14]. ‘‘We failed and failed and failed, until a couple years ago suddenly we got DNA out. We had a breakthrough, and we could basically see this whole ecosystem come to life.” In another few years, he said, more advances could allow even more information to be extracted from the same samples.













 京公网安备 11010502051620号
京公网安备 11010502051620号




