《1. Introduction》
1. Introduction
A commercial herbicide with a clearly new molecular target (site of action (SOA)) has not been introduced for more than 30 years [1]. During this period, the number of unique cases of evolved herbicide-resistant weeds has increased by about 500%, with many of the most problematic weed species now having resistance to several herbicides with different SOAs [2]. One approach to managing the risk of evolution of herbicide resistance is to apply herbicides with several modes of action (MOAs) in combination, just as pharmaceutical mixtures delay evolution of resistance to drugs. As with antibiotics and some other pharmaceuticals (e.g., anticancer drugs), there is a dire need of compounds with new SOAs for resistance management.
Unfortunately, the agrochemical industry’s investment in herbicide discovery declined in the mid-1990s due to the huge success of glyphosate-resistant crops and the resulting decrease in the value of the herbicide market [1]. The rapid consolidation of companies involved in herbicide discovery [3] also contributed to the decline in overall investment in herbicide discovery. The dire need for discovery of herbicides with new SOAs has fed the interest in approaches to achieving this goal. Historically, discovery programs have relied on the synthesis and evaluation of large chemical libraries to identify leads with promising herbicidal activity (high-throughput screening). Another approach has been to optimize the structure of a moderately active compound with a novel molecular target to develop commercial products with improved activity at the molecular target and physicochemical properties needed for agricultural use. Examples of this are the triketone herbicides that were derived via structural optimization of the allelochemical leptospermone [4]. Leptospermone and these commercial herbicide analogs inhibit hydroxyphenylpyruvate dioxygenase (HPPD) [5], the last new herbicide SOA introduced. Many other natural phytotoxins with novel molecular targets exist that could be used as starting templates for herbicide discovery [6]. However, this approach has not yielded a commercial herbicide with a new SOA to date. Likewise, neither in silico modeling of small molecules binding to potential herbicide target enzymes (biorational design) or screening large combinatorial chemical libraries with in vitro assays of potential target enzymes have resulted in commercial products with new SOAs. Lastly, using gene knockouts to search for new SOAs has identified potential new herbicide targets (discussed in detail in Ref. [7]), but finding molecules for these target sites with good herbicidal activities has not been successful. One problem with this approach is that knockout mutants generally eliminate activity of the target enzyme completely, whereas chemicals rarely provide 100% inhibition of the enzyme. Thus, a lethal phenotype achieved with a knockout does not translate into herbicidal activity with partial inhibition of the target’s activity by a chemical. A calibrated method of gene knockouts is needed to better evaluate herbicide SOAs.
Despite a strong interest in novel approaches to discovery of herbicides with new SOAs, there are few papers that provide insight into new strategies for such discoveries. An exception is the use of genomic approaches to discover target enzymes of phytotoxins from genes that are located in gene clusters encoding the enzymes of biosynthetic pathways for natural phytotoxins (e.g., Ref. [8]). Microbes that produce toxins that inhibit enzymes of primary metabolism often have a gene for resistance in the gene cluster for the toxin synthesis [9]. In some cases, the resistance gene encodes a mutant, resistant form of the target of the toxin. Thus, study of microbial phytotoxins has the potential for discovery both new potential herbicides and their SOAs. This approach is promising, but unproven. For example, although a microbially-produced natural product inhibitor (aspterric acid) of the branched chain amino acid synthesis pathway enzyme dihydroxy acid dehydratase (DHAD) was discovered by this method, it is weak inhibitor of the enzyme and a weak herbicide [10]. Although DHAD has been considered a potential herbicide SOA for some time (discussed in Ref. [9]), there are still no commercial herbicides that inhibit DHAD.
In this paper, two new potential approaches to herbicide target site identification are discussed. The first is identification of enzymes that, when inhibited, would cause accumulation of phytotoxic metabolites (Fig. 1). The second is identification of target sites with low molecular concentrations in plant tissues (Fig. 1). Both of these approaches are likely to identity targets that respond to relatively low herbicide doses. Low doses are desirable from environmental and toxicological standpoints, and, if a molecule is expensive to produce, a low dose can make it economically feasible as a herbicide.
《2. Toxic precursors of primary metabolism enzymes》
2. Toxic precursors of primary metabolism enzymes
Most commercial herbicides have only one primary molecular target site in weeds. In some cases, the herbicide targets several variations of the same enzyme type, as with the serine–threonine protein phosphatase inhibitor endothall and herbicides that inhibit very long chain fatty acid synthases. Almost all of these targets are enzymes of primary metabolism. The herbicides with non-enzyme targets are the photosystem II inhibitors of photosynthetic electron flow (e.g., atrazine and diuron), photosystem I energy diverters (paraquat and diquat), and the auxinic herbicides (e.g., 2,4- dichlorophenoxyacetic acid (2,4-D)) that bind F-box proteins, which are not enzymes, but mediate signal transduction and gene expression.
Blocking a primary metabolism pathway by inhibition of an enzyme should be lethal, however, complete blockage with a chemical is difficult. The accumulation of a phytotoxic compound, in addition to partial blockage of the pathway should increase the efficacy of a herbicide. Several highly successful herbicides and some natural phytotoxins exert their effects largely due to accumulation of phytotoxic intermediate metabolites or metabolite derivatives (Fig. 1). The clearest case of this is that of the protoporphyrinogen oxidase (PPO) inhibitors (Table 1) [11]. PPO inhibitors are a relatively large class of herbicides. PPO is an enzyme found in both mitochondria and plastids (e.g., chloroplasts) involved in biosynthesis of porphyrins needed for heme and chlorophyll production. These inhibitors cause the accumulation of the enzymatic product of PPO, protoporphyrin IX (PPIX), even though they inhibit PPO. PPIX is highly toxic in the presence of light and molecular oxygen, as under these circumstances it acts as a photosensitizing pigment, generating singlet oxygen, which in turn produces other reactive oxygen species (ROS). When PPO is inhibited, its precursor, protoporphyrinogen IX, accumulates and exits the porphyrin pathway to be oxidized to PPIX by means other than PPO in parts of the cell that are relatively unprotected from singlet oxygen and other ROS. Normally, PPIX and other porphyrin pathway intermediates are found in very low concentrations and are confined to the porphyrin pathway in mitochondria and plastids, where there is relatively good protection from ROS, especially in the chloroplast. An advantage of this type of MOA is that lethal ROS levels are probably produced by partial inhibition of the PPO activity in the plant. The most potent PPO inhibitors are some of most active herbicides with the lowest application rates (a few grams per hectare). At lethal doses, all herbicides cause generation of ROS as a tertiary effect of severe biochemical and physiological disruption. The ROS further disrupt the plant’s biochemistry and physiology. There are many articles that mistake this universal tertiary effect with a more primary effect. With PPO inhibitors, the generation of ROS is much closer to the primary event of PPO inhibition.
《Fig. 1》
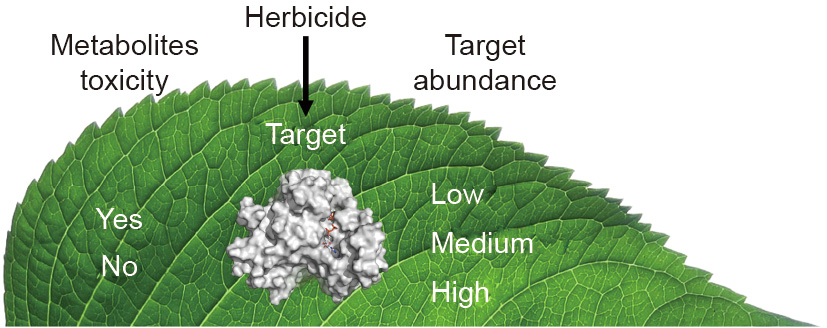
Fig. 1. Illustration of the two new potential approaches to herbicide target site identification discussed in this paper. The first is identification of enzymes that, when inhibited, would cause accumulation of phytotoxic metabolites (left), and the second is identification of target sites with low molecular concentrations in plant tissues (right).
《Table 1》
Table 1 Herbicide MOAs that involve accumulation of toxic precursors and/or involve target sites (SOAs) with low concentrations.
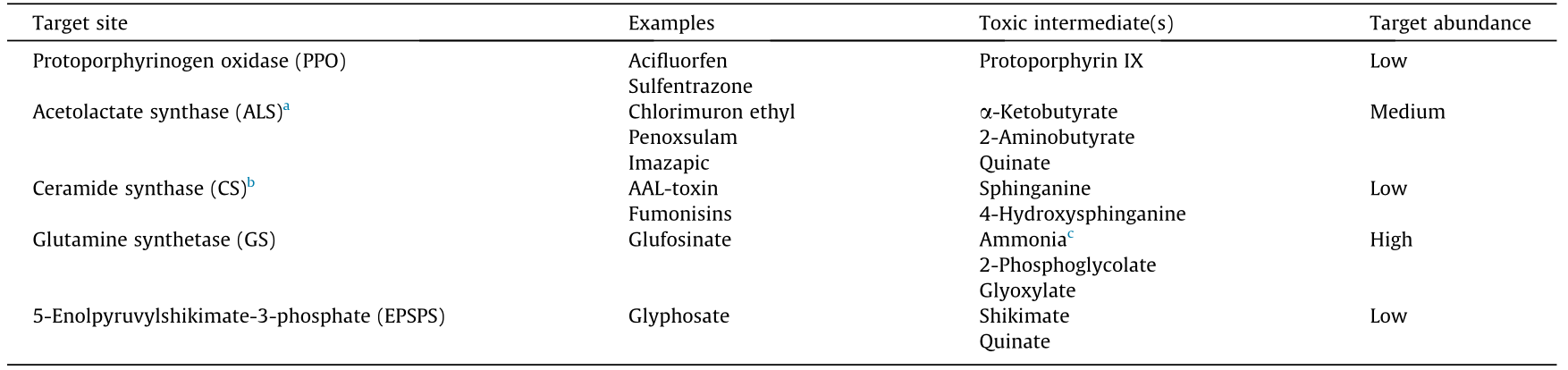
AAL: Alternaria alternata f. sp. lycopersici.
a Also called acetohydroxy acid synthase (AHAS).
b Also called sphinganine N-acyltransferase.
c Glufosinate causes accumulation of ammonia but this may not be related to its herbicidal activity.
Acetolactate synthase (ALS) inhibitors are a very large class of herbicides (Table 1). ALS is the first enzyme committed to the synthesis of branched chain amino acids (valine, leucine, and isoleucine). If the action of ALS inhibitors is due entirely to depletion of branched chain amino acids, theoretically, either of the other two enzymes of this pathway could be equally good herbicide targets. Yet, there are many commercial ALS inhibitor herbicides from several chemical classes, and there are no herbicides that target either of the other two enzymes of the pathway. Good in vitro inhibitors of keto acid reductoisomerase (KARI) and DHAD, the second and third enzymes in the pathway, have been found [12–14]. But, efforts to develop commercial herbicides from both KARI and DHAD inhibitors have been unsuccessful. One difference between these three enzymes is that only the ALS substrate, α-ketobutyrate (2KB), is phytotoxic. 2KB can be transaminated to phytotoxic 2- aminobutyrate (2AB), and both 2KB and 2AB accumulate in higher plants treated with ALS inhibitors [15]. Although Shaner and Singh [16] concluded that since there is no good correlation between 2AB and the herbicidal effect of an ALS inhibitor, lack of a good correlation does not preclude lack of involvement. In the same paper, they showed that treatment of maize seedlings with high levels of valine caused growth inhibition and accumulation of 2AB. Since 2AB is phytotoxic, this may explain the growth reduction by valine. The lack of good correlations in their experiments could have been due to confounding effects of the cellular localization of the 2AB pools involved in phytotoxicity. ALS inhibitors also cause accumulation of quinate, a phytotoxin from the shikimate pathway, as discussed below [17]. How inhibition of ALS leads to quinate accumulation is unclear. The best ALS inhibitors are herbicidal at very low doses, compared to most other commercial herbicides.
Alternaria alternata f. sp. lycopersici (AAL)-toxin is a natural phytotoxin that is effective at very low doses. By inhibition of ceramide synthase (CS or sphinganine N-acyltransferase in plants), it causes rapid and high levels of accumulation of the sphingoid base CS precursors sphinganine and 4-hydroxysphinganine (Table 1) [18]. The effects are rapid and strong, even at sub-micromolar concentrations. Furthermore, exogenously supplied CS precursors cause phytotoxicity symptoms similar to those of AAL-toxin [19], indicating that the accumulation of precursors, rather than inhibited production of ceramides is the cause of the rapid loss of membrane function. The levels of CS precursors in healthy plant tissues are very low, but ceramide and its glucosylated forms are significant components of plant plasma membranes [20]. The very rapid effect of free sphingolipid bases on loss of membrane integrity [19] is probably due to direct effects of these compounds on plasma membrane integrity, rather than indirect effects from another mechanism. AAL-toxin and other structurally related CS inhibitors (e.g., fumonisins) cause similar effects on accumulation of CS precursors [21]. Although many have invoked induction of apoptosis as the MOA of AAL toxin (e.g., Ref. [22]), these effects are clearly secondary or tertiary to the primary effects of the toxin on CS, just as the apoptosis associated with paraquat toxicity in plants [23] is clearly a secondary effect of the herbicide. In full sunlight, the very rapid membrane destruction caused by massive ROS production, resulting in cellular death caused by paraquat is too rapid to involve apoptotic processes.
Glufosinate is a broad-spectrum and non-selective herbicide that causes rapid plant death (contact activity). Glufosinate is a racemic mixture of D- and L-phosphinothricin. L-Phosphinothricin is a natural product ot Streptomyces hygroscopus, and this enantiomer is the only active component of glufosinate. It is used for weed management in non-crop areas and in transgenic glufosinate-resistant crops. It inhibits glutamine synthetase (GS), an enzyme present in high abundance in plant leaves, where it plays a vital role in plant nitrogen assimilation (Table 1) [24]. The GS1 isoform is localized in the cytosol, whereas the GS2 isoform is found in chloroplasts [25]. GS2 assimilates ammonia generated by photorespiration into glutamine [26,27]. While there are several known GS inhibitors, mostly compounds produced by microbes, glufosinate is the only molecule to have been developed into a commercial herbicide [6].
Glufosinate binds to GS irreversibly and results in accumulation of ammonia derived from the photorespiratory pathway. While there is an association between the mechanism of action of glufosinate and its alteration of the photorespiration pathway, the toxic effect of glufosinate is not directly linked to ammonia accumulation but instead the results of rapid accumulation of ROS and subsequent lipid peroxidation [28]. The origin of the ROS is not well understood but could be related to inhibition of carbon assimilation observed in sensitive plants treated with this herbicide. This inhibition is likely due to the accumulation of some of the intermediate in the photorespiratory pathway (phosphoglycolate, glycolate and glyoxylate). Some of these intermediates are strong inhibitors of ribulose-1,5-biphosphate carboxylase/oxygenase (RUBP carboxylase or Rubisco) [29,30], and could thus be considered natural phytotoxins.
Glyphosate is the world’s most used herbicide [31]. It has only one SOA, 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS), but how inhibition of EPSPS kills plants (the MOA) is still not entirely clear (Table 1) [32]. EPSPS is an enzyme of the shikimate pathway, which is responsible for production of aromatic amino acids (phenylalanine, tyrosine, and tryptophan). Most plant species have very low levels of shikimate-3-phospate (the substrate of EPSPS) or shikimate (the substrate of the enzyme just before EPSPS), but treatment with glyphosate causes high levels of accumulation of shikimate and to a lesser extent quinate, another product of an intermediate (3-dehydroquinate) of the shikimate pathway. Quinate can also be generated from shikimate by quinate hydrolase [33]. Interestingly, ALS inhibitor herbicides (see above) also cause accumulation of quinate [17]. Plants that are resistant to glyphosate by any resistance mechanism(s) do not accumulate shikimate, so measurement of shikimate in response to glyphosate is a quick bioassay to detect glyphosate resistance [34]. We have found no data on phytotoxicity of shikimate, but quinate is moderately phytotoxic, perhaps causing some of the effects of ALS inhibitors and glyphosate [17,35,36]. Some of the effects of quinate, such as effects on carbohydrate metabolism, are similar to those of glyphosate [35]. Thus, at least part of the effects of glyphosate are probably due to high levels of quinate.
Although both shikimate and quinate are found at very low levels in most plant tissues, there are a few plant species that accumulate high levels of shikimate (e.g., star anise (Illicium verum), sweetgum (Liquidamber styraciflua)) [37,38], and quinate (e.g., Chinchona officianalis) [39]. We assume that such plants have a means of compartmentalizing these compounds away from cells involved in normal growth and development in order to avoid autotoxicity, as is commonly found with many other compounds that can cause autotoxicity to plants [40].
Many years ago, Cornish-Brown predicted that pesticides that act by uncompetitive inhibition were likely to be especially effective because of strong accumulation of metabolic intermediates [41]. Such an effect would be magnified if the intermediates were toxic to the target organism. Glyphosate is the only commercial herbicide known to be an uncompetitive (with shikimate) enzyme inhibitor, and it causes large accumulation of two intermediates. Thus, uncompetitive enzyme inhibitor herbicides that inhibit an enzyme with phytotoxic precursors might be especially effective.
There are many phytotoxic metabolites of secondary metabolism, but the enzymes that produce them are not likely to be good herbicide targets. This is for several reasons. First, if secondary products are highly phytotoxic, they are compartmentalized or excreted to avoid autotoxicity. For example, the highly phytotoxic artemisinin is compartmentalized in the subcuticular space of glandular trichomes, away from cytoplasm [40,42]. Some phytotoxic compounds are stored in the vacuole, where they can do no harm. For example, phytotoxic glucosinolates are stored in plant vacuoles [43]. The hydroquinone precursor of the allelochemical sorgoleone is secreted by root hairs of Sorghum species into soil, where it oxidizes to the phytotoxic quinone sorgoleone [44]. These secondary compounds are usually at the end of biosynthetic pathways, so they are not substrates that could accumulate by inhibiting an enzyme. Another problem is that the most phytotoxic secondary metabolites of plants are produced by a very limited number of plant species, most of which are not likely to be target weeds. An extreme example of this is artemisinin, which is produced only by Artemisia annua L., a very minor weed [42]. There are secondary compounds that could almost be considered primary metabolites, as they are virtually ubiquitous in higher plants. An example is t-chalcone, a precursor for phenylpropenoids in higher plants. It is a moderately effective phytotoxin [45], so inhibition of the enzyme that uses it as a precursor, chalcone isomerase, could cause toxic levels of it to accumulate.
Another factor that might enhance the production of phytotoxic intermediates is deregulation of the metabolic pathway when the SOA is inhibited. This is something of which we know very little. There is some evidence that inhibition of EPSPS results in lower concentrations of one or more shikimate pathway products that regulate carbon flow into the shikimate pathway [32]. Something similar could be happening with inhibition of ALS, CS, GS, and PPO. If so, the level of pathway deregulation could be critical as to whether the concentration of toxic intermediate rises to a lethal level.
Another factor to consider is the in vivo half-life of the phytotoxic intermediate. The compound could be unstable or might be metabolically converted to a less or more toxic compound than the intermediate. An example of the latter is the conversion of shikimate to hydroxybenzoic acids such as protocatechuic and gallic acids [46]. Gallic acid is a phytotoxin [47].
These cases of inhibition of an enzyme causing accumulation of toxic metabolites may represent a small fraction of the possibilities along these lines. To our knowledge, there has been no study to examine the phytotoxicity of all primary metabolites, much less secondary metabolites, to plants. Another approach might be to examine metabolomic data to find which compounds are present in very low concentrations in healthy plant tissues, as evolution has minimized accumulation of phytotoxic primary metabolic intermediates in plants. These compounds can then be tested for phytotoxicity to identify potential herbicide target enzymes (SOAs).
《3. Low concentration of targets》
3. Low concentration of targets
Another approach to viable herbicide target identification is to determine those targets which are present at relatively low concentrations in plants (Fig. 1). Conversely, it has long been recognized that potential target sites that are present in great abundance are not good target sites because of the high doses of herbicides that would be required to inhibit a sufficient fraction of the target to kill the plant. An example of this is ribulose-1,5- bisphosphate carboxylase/oxygenase (Rubisco), the enzyme responsible for C3 photosynthetic carbon fixation. Rubisco is the most abundant protein in plants, making up about 50% and 30% of the protein in green leaves of C3 and C4 plants, respectively [48]. Because of its abundance, it has even been proposed as a source of dietary protein from green leaves [49]. There are both natural and synthetic inhibitors of Rubisco, such as 2-carboxyarabinitol-1-phosphate, a naturally occurring transition state intermediate of the Rubisco intermediate [50], and iodoacetol, a synthetic compound [51], but to inhibit sufficient Rubisco to kill weeds would take massive application rates. Thus, this is not a target site in which those involved in herbicide discovery have any interest.
The case of evolution of resistance to glyphosate by gene amplification of its target, EPSPS, is proof that the amount of the target site of herbicide can be critical to whether the herbicide is viable. Glyphosate is a high use rate herbicide, requiring ~1 kg·hm-2 to effectively kill most of the target weed species in many situations. Being the most used herbicide on earth [31], the selection pressure to evolve resistance has been enormous, with 47 weed species having become resistant in just over 20 years, beginning about 25 years after commercialization of glyphosate [2]. Various mechanisms of resistance evolved [52,53], but one of the more commonly found mechanisms is amplification of a gene for EPSPS, resulting in a much higher concentration of the EPSPS enzyme in the weed, thus requiring a much higher dose of glyphosate to inhibit enough EPSPS to lethally block the shikimate pathway [54]. A 90-fold increase in the copies of the gene led to a 12-fold increase in the amount of EPSPS protein in glyphosate-resistant Amaranthus palmeri [54], an amount that results in about a six- to eight-fold increase in the effective dose of glyphosate, a dose that is neither economically nor environmentally viable [54]. Since this first case of gene amplification, this mechanism of resistance to glyphosate has been found in both broadleaf and grass species (e.g., Refs. [54–59]). If the levels of EPSPS in weeds were normally as high as those with evolved gene amplification of the gene for this herbicide target, glyphosate would have never been developed as a herbicide, as it would require at least 10 kg of herbicide per hectare, a dose that would be economically and environmentally unacceptable. If it were a low use rate herbicide, this might not be the case. Gene amplification has been found as a mechanism of resistance to acetyl-coenzyme A (CoA) carboxylase inhibitor herbicides also [60]. This mechanism of resistance has not been looked for in most cases of herbicide resistance or in cases of natural tolerance of some species to certain herbicides, so we do not fully know how important the role of enzyme abundance is in many cases of evolved resistance and natural tolerance.
Likewise, there is almost no information available on the amount (either absolute or relative) of protein found in plants for each enzyme of primary biochemical pathways. This amount represents the number of potential herbicide binding sites and, as with glyphosate, it will influence the concentration of herbicide that must arrive at the subcellular sites of these enzymes to be lethal to a plant. Proteomics has made great advances in the past two decades, providing a huge amount of information on comparative proteomics, subcellular location of proteins, protein function, post-translational modifications, etc., but, even for Arabidopsis thaliana, the actual relative concentrations of each protein of all enzymes of primary metabolism have not been determined. However, methods are available for the identification of low abundance proteins in plant cells to levels as low as 2.25 fmol·mg-1 leaf fresh weight (e.g., Ref. [61]). Low abundance enzymes are more likely to be better potential herbicide target sites than high abundance enzymes.
《4. Parting thoughts》
4. Parting thoughts
There are caveats regarding both of these potential approaches related the fact that pool size or concentration does not reflect turnover rate of the pool. In the case of a toxic metabolite, a higher turnover rate of the pool would mean a more rapid accumulation of the compound when the complementary SOA is inhibited, increasing the effect of the herbicide. In the case of an enzyme SOA, a more rapid turnover rate might reduce the efficacy of the herbicide targeting that SOA, as the potential for interaction of the herbicide with the target in a unit of time would go up, as more copies of the enzyme would exist in a unit of time. This would be more important if the herbicide is an irreversible binder, and degradation of the enzyme does not release an unaltered herbicide. The fate of the inhibited enzyme and/or the herbicide binding the enzyme would affect the influence of enzyme pool flux rate on herbicide efficacy. These caveats must be kept in mind.
The urgent need for commercial herbicides with new SOAs makes it worthwhile to use all discovery strategies that may maximize the likelihood of success. In this short paper, we have proposed two strategies that are conceptually simple. Metabolomics can be used to identify targets that might have toxic precursors or precursors that might be converted to a toxin if accumulating in vivo. Proteomics can be used to identify enzyme targets present in low abundance. Ideally, SOAs in low abundance that produce phytotoxins when inhibited, such as PPO (Table 1) should be sought. However, this is not always the case, as demonstrated by glufosinate targeting GS, the second most abundant enzyme in leaves. It is possible that both strategies that we have discussed have been used by a herbicide discovery company, but, if so, we have not found a published record of it.
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Franck E. Dayan and Stephen O. Duke declare that they have no conflict of interest or financial conflicts to disclose.














 京公网安备 11010502051620号
京公网安备 11010502051620号




