《1. Introduction》
1. Introduction
Ochrobactrum anthropi (O. anthropi ) is a Gram-negative Bacillus that is tremendously abundant in the environment. It was formerly placed in the category of CDC group Vd [1], which belongs to the family Brucellaceae [2]. O. anthropi was previously considered to be a human conditional pathogen that mainly infected immunocompromised patients [1,3,4]. However, several reports have indicated that O. anthropi can also cause infections in healthy humans [5–7]. The manifestations of O. anthropi infections are endophthalmitis [6], endocarditis, meningitis, osteochondritis, osteomyelitis, pancreatic abscess, puncture wound, pelvic abscess, and urinary tract infection [8–14]. Besides human beings, O. anthropi has been reported to infect other mammals. Franci et al. [15] were the first to report a healthy canine being infected with O. anthropi; this occurred after routine dental cleaning as a result of the anesthetic solution being contaminated with O. anthropi. El-Adawy et al. [16] separated O. anthropi from the cecum of a turkey. To date, O. anthropi has not been reported as occurring in any other species. In this study, we isolated and characterized one strain of O. anthropi from a pig for the first time.
《2. Materials and methods》
2. Materials and methods
《2.1. Isolation of bacteria from pig clinical samples》
2.1. Isolation of bacteria from pig clinical samples
The pig brain sample was obtained from a commercial farm in Jiangxi Province, China. The pig had neurological symptoms, and the pathological examination showed meningitis at necropsy. Histopathological examination and bacterial separation were carried out from the brain samples. Swabs were taken from the brain sample under aseptic conditions to avoid contamination. The isolate was cultured on tryptone soy agar (TSA; BD DifcoTM, Becton, Dickinson and Company, USA) containing 10% newborn bovine serum (Zhejiang Tianhang Biotechnology Co., Ltd., China) at 37 °C for 24 h. Gram staining was conducted according to the method described by Gerhardt et al. [17]. The biochemical reaction was profiled using the API 20 NE assay (bioMérieux Inc., France), according to the manufacturer’s instruction.
《2.2. DNA preparation》
2.2. DNA preparation
DNA from bacterial samples cultured for 24 h was extracted by a GENEWIZ Bacteria Genomic DNA Kit (GENEWIZ, China) according to the manufacturer’s instruction.
《2.3. PCR, gene sequencing, and construction of phylogenetic trees》
2.3. PCR, gene sequencing, and construction of phylogenetic trees
The 16S rRNA gene and recA gene were amplified using the methods previously described by Scholz et al. [18]. The forward and reverse primers 27f (5' -AGAGTTTGATCMTGGCTCAG-3' ) and 1492r (5' -AAGTCGTAACAAGGTARCCG-3' ) were used to generate an approximately 1400 bp fragment of the 16S rRNA gene. The profile of the polymerase chain reaction (PCR) cycle was as follows: 94 °C for 5 min, 30 cycles at 94 °C for 60 s, 56 °C for 30 s, 72 °C for 90 s, and 10 min at 72 °C. The PCR products were analyzed in 1% agarose by gel electrophoresis. The primers for recA are recA-f (5' -ATGTCTCAAAATTCATTGCGAC-3' ) and recA-r (5' -AGCATCTTCTTCCGGTCCGC-3' ); these were used to obtain a 1065 bp size band of the recA gene. The PCR cycle was as follows: 94 °C for 5 min, 30 cycles at 94 °C for 30 s, 58 °C for 30 s, 72 °C for 60 s, and 10 min at 72 °C. The PCR products were analyzed by electrophoresis in 1% agarose gel.
The PCR products were sent to GENEWIZ for sequencing. The sequences were BLASTed (where BLAST stands for Basic Local Alignment Search Tool) by BLASTnx in the National Center for Biotechnology Information (NCBI) database. CLUSTAL X was used [19] to align the sequences. Phylogenetic trees were built based on the 16S rRNA gene ( rrs ) and the recA gene using the neighborjoining method in Kimura’s two-parameter model [20] in MEGA Version 5 [21,22]. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test (1000 replicates) were shown next to the branches [23].
《2.4. Genomic sequencing and BLAST》
2.4. Genomic sequencing and BLAST
The genomic sequences were BLASTed by BLAST genomes in the NCBI database. Partial sequences of the recA and 16S rRNA and the genomic sequences Ochrobactrum and Brucella spp. were deposited in GenBank with the accession numbers documented in Table S1 in Appendix A. Supplementary data.
《2.5. Antibiotic susceptibility and resistance testing》
2.5. Antibiotic susceptibility and resistance testing
The isolate was subjected to antibiotic susceptibility and resistance testing by the disk diffusion method presented by Bauer et al. [24]. The isolated strain was cultured on TSA containing 10% newborn bovine serum at 37 °C for 24 h and was then diluted in phosphate-buffered saline (PBS) (0.01 mol·L-1 , pH 7.2) at 1 × 108 CFU·mL-1 (where CFU stands for colony forming units). The bacterial suspension was spread on the TSA containing 10% newborn bovine serum with a sterile cotton swab, and the paper antibiotic disks (Hangzhou Microbial Reagent Co., Ltd., China) were placed on the inoculated agar surface. The plates were incubated at 35 °C for 24 h prior to determination. The zone diameters of each drug were interpreted using the criteria published by the Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria from Animals [25].
《2.6. Mouse experimental infection study》
2.6. Mouse experimental infection study
The isolated strain was cultured on TSA containing 10% newborn bovine serum at 37 °C for 24 h and was then diluted in PBS (0.01 mol·L-1 , pH 7.2) to obtain a series suspension of about 5 × 109 –2 × 1010 CFU·mL-1 . BALB/c mice were intraperitoneally injected with the bacterial solution at 1 × 109 –4 × 109 CFU per mouse with a volume of 0.2 mL per mouse. Histopathological observation was carried out for the mouse brain samples. The animal trial followed the animal protocol approved by the Animal Care and Ethics Committee of the China National Research Center for Veterinary Medicine.
《2.7. Histopathological observation》
2.7. Histopathological observation
The brain samples were fixed with 10% buffered formalin overnight, followed by embedding in paraffin. Brain tissues were sectioned and stained with hematoxylin and eosin for histopathological evaluation.
《3. Results》
3. Results
《3.1. Histopathological examination and bacterial isolation》
3.1. Histopathological examination and bacterial isolation
The histopathological results for the pig brain sample showed necrotic nodules in the white matter of the brain, necrosis of gray matter neurons, and a large number of neutrophil infiltrations, as shown in Fig. 1. The brain sample was streaked onto TSA plates to obtain pure colonies. The colonies were small and smooth; the cells in the colonies were shown to be Gram-negative bacillus after Gram staining (Fig. 2).
《Fig. 1》

Fig. 1. Histopathological observation of the pig brain sample (hematoxylin and eosin staining (H & E), 20×). (a) Necrotic nodules in the white matter of the brain (shown by the arrow); (b) necrosis of gray matter neurons (shown by the triangle) and neutrophil infiltration (shown by the arrow).
《Fig. 2》

Fig. 2. Colony morphology and cell morphology of the isolate. (a) Colony morphology of the isolate; (b) microscopic examination of cell morphology (100×) after Gram staining.
Biochemical identification revealed that the isolated strain was oxidase and catalase positive. The isolate was identified as O. anthropi (99.9% probability) by API 20 NE test (Table 1). This isolate was named JXLH03B.
《Table 1》
Table 1 API 20 NE identification of the JXLH03B isolate.


《3.2. Sequencing and phylogenetic analysis》
3.2. Sequencing and phylogenetic analysis
The sequence of the JXLH03B 16S rRNA gene was completely identical to that of Ochrobactrum sp. (GenBank accession No. MH201346.1), O. anthropi (GenBank accession No. LT671861.1), and Ochrobactrum lupini (O. lupini) (GenBank accession No. KU217325.1). The recA gene was also BLASTed and was shown to be completely identical to O. anthropi (GenBank accession No. LT671861.1).
Phylogenetic analysis was performed based on the 16S rRNA gene. As shown in Fig. 3, JXLH03B was grouped with O. anthropi LMG 7991, O. anthropi OAB, O. anthropi LMG 5140, O. anthropi LMG 3333, O. anthropi LMG 3333T, O. anthropi DSM 14396, O. anthropi ATCC 49188, O. anthropi 7b2C, and O. lupini LMG 1821. By contrast, JXLH03B was grouped with O. anthropi DSM 14396, O. anthropi LMG 3333, and O. anthropi 10CS0094, based on the recA gene (Fig. 4).
《Fig. 3》

Fig. 3. Phylogenetic analysis based on the 16S rRNA gene using the CLUSTREE neighbor-joining method. Scale bar: 0.002 divergent residues per site. The significance of each branch is indicated by a bootstrap value calculated for 1000 subsets.
《Fig. 4》
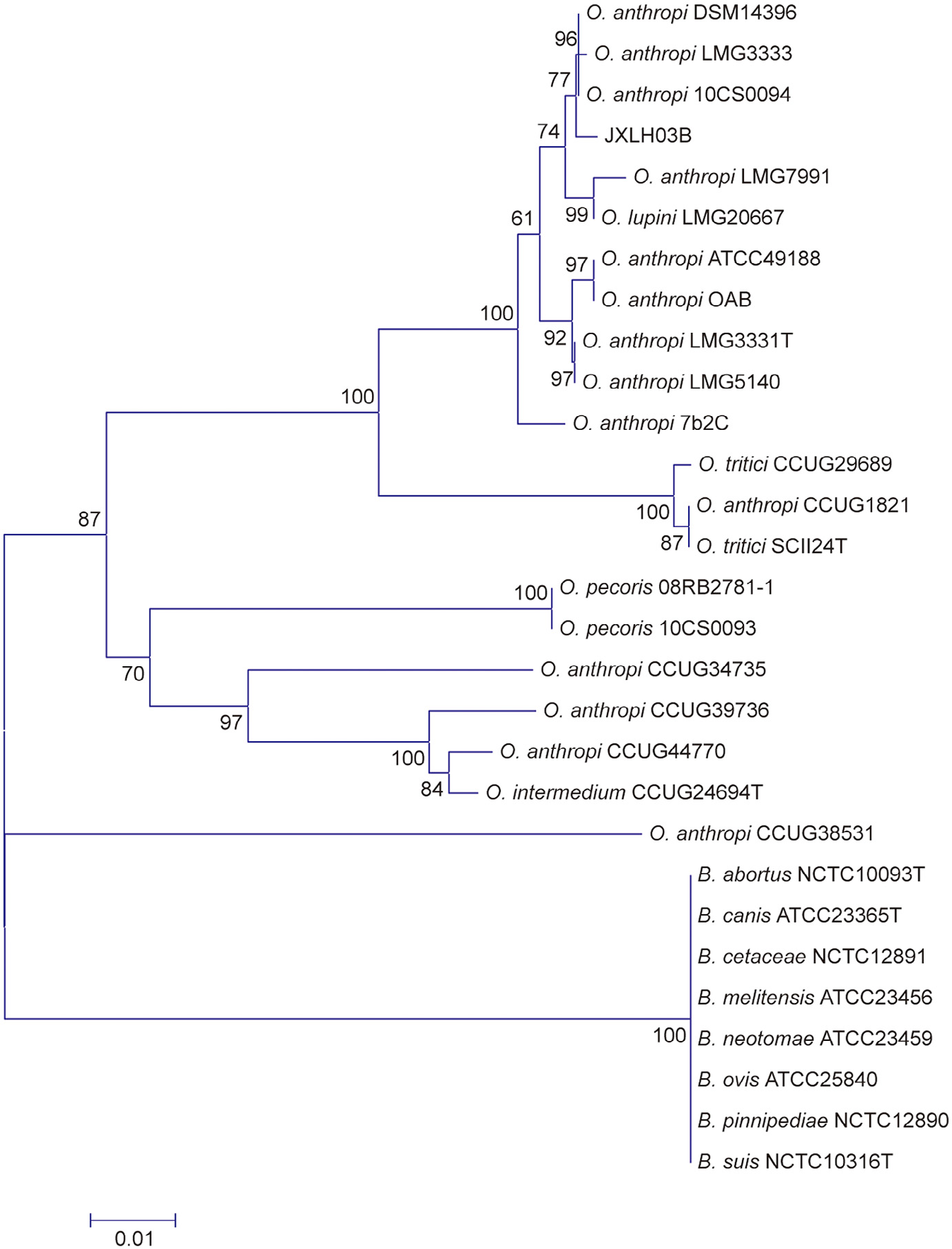
Fig. 4. Phylogenetic analysis based on the recA gene using the CLUSTREE neighbor-joining method. Scale bar: 0.01 divergent residues per site. The significance of each branch is indicated by a bootstrap value calculated for 1000 subsets.
《3.3. Genomic sequencing and BLAST》
3.3. Genomic sequencing and BLAST
The genomic sequencing results of the JXLH03B isolate were two sequences with a total length of 4 725 913 bp. The G + C content was 56.01%. The lengths of sequence 1 and sequence 2 were 2 641 072 bp and 2 084 841 bp, respectively. The BLAST results showed that the similarity between the JXLH03B isolate sequences and the two deposited genome sequences of O. anthropi was 97.54%–98.08% (Table 2).
《Table 2》
Table 2 Sequence similarity of genomic sequences between JXLH03B and two different O. anthropi strains.

《3.4. Mouse experimental infection study》
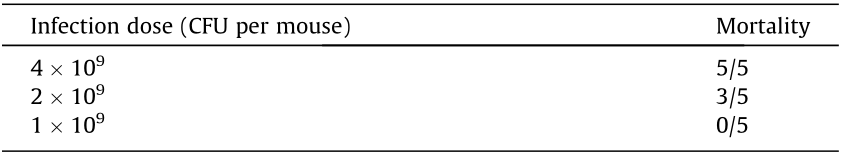
3.4. Mouse experimental infection study
BALB/c mice were infected with JXLH03B at 1 × 109 – 4 × 109 CFU per mouse. Mouse mortality ranged from 0/5 to 5/5 (Table 3). The brains of all the dead mice were sampled for histopathological examination. All of the mice that died after infection had inflammation in the brain tissue. The histopathological results showed several necrotic foci in the brain. The brain parenchyma was liquefied, and a large amount of neutrophils necrosis aggregated to form nodules (shown by the arrow in Fig. 5). Some glial cells were found to collapse (Fig. 5).
《Table 3》
Table 3 Results from mice infected with JXLH03B.
《Fig. 5》
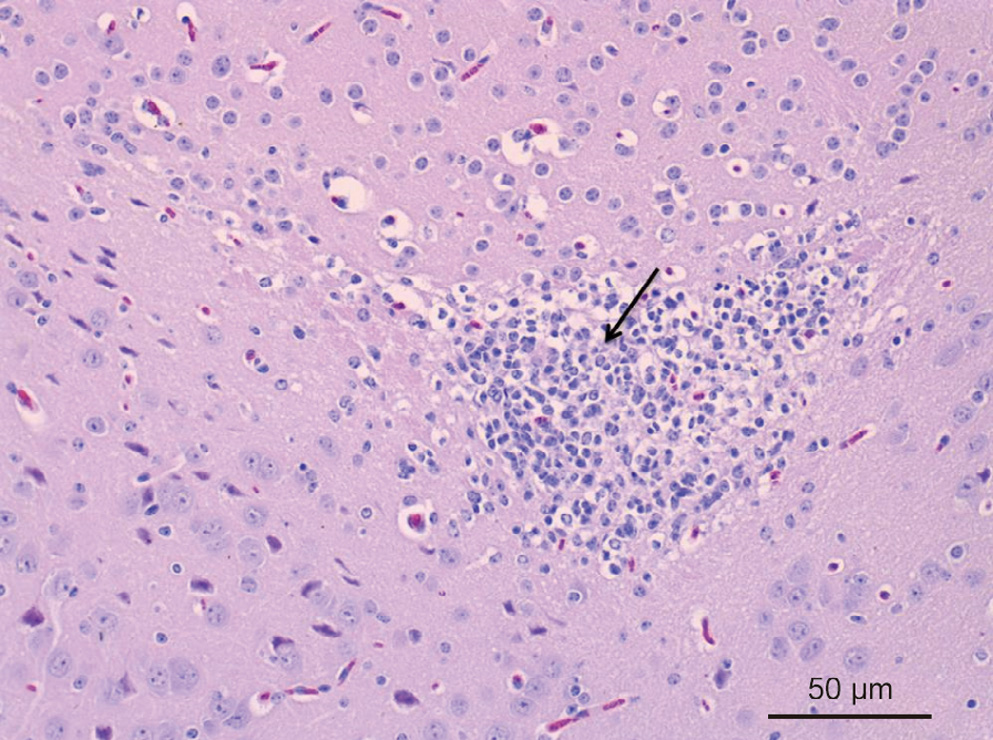
Fig. 5. Histopathological observation of the mouse brain sample (H & E, 20×). Necrotic foci occurred in the brain, and neutrophils necrosis aggregated to form nodules (shown by the arrow).
《3.5. Antibiotic sensitivity test》
3.5. Antibiotic sensitivity test
Antimicrobial susceptibility testing was performed using a disk diffusion test following the guidelines of the Performance Standards for Antimicrobial Disk and Dilution Susceptibility Tests for Bacteria from Animals [25]. The JXLH03B isolate was susceptible to gentamicin sulfate, amikacin, enrofloxacin, ciprofloxacin, doxycycline, and florfenicol, but was resistant to penicillin sodium, ceftiofur, kanamycin sulfate, lincomycin, and tylosin (Table 4).
《Table 4》
Table 4 Disk diffusion test values and interpretation of the JXLH03B isolate.
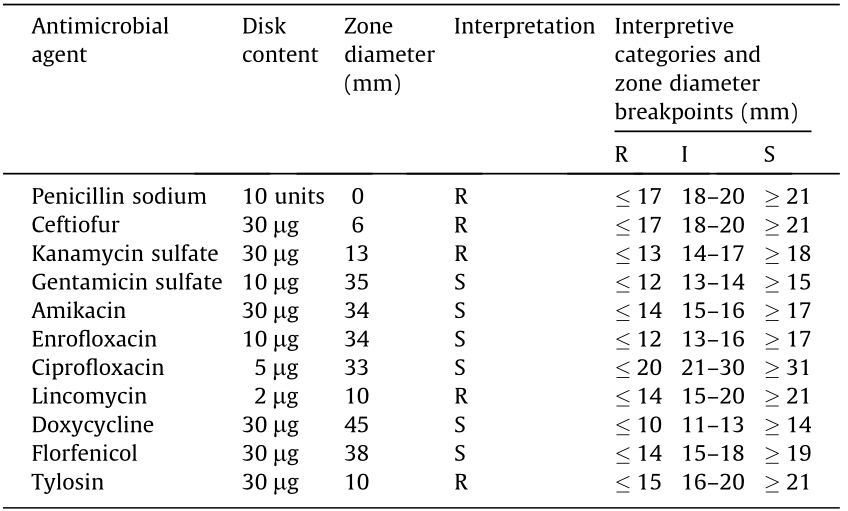
S: sensitive, R: resistant, I: intermediate.
《4. Discussion》
4. Discussion
O. anthropi is an aerobic, non-lactose-fermenting, motile, oxidase-positive, urease-positive, Gram-negative Bacillus previously known as “Achromobacter group Vd” [1]. The biochemical reaction profiles (API 20 NE) identified the JXLH03B strain as O. anthropi. The genus Ochrobactrum belongs to the family Brucellaceae and has a close phylogenetic relatedness to Brucella [26]; the genus Ochrobactrum comprises about 16 species [27]. 16S rRNA gene analysis was used for the identification and differentiation of Ochrobactrum and Brucella spp. [2,26,27]. However, this approach—particularly partial sequencing—is prone to misidentifying these pathogens because of their high sequence similarities [27]. Meanwhile, recA analysis provides more accurate identification and differentiation [18,27]. We performed the 16S RNA gene sequencing alignment of the JXLH03B isolate, which could not be completely identified as O. anthropi; however, the result of the recA gene sequencing comparison identified the JXLH03B isolate as O. anthropi.
The infection test in mice showed that the strain had a certain pathogenicity that led to brain lesions. To test the pathogenicity of the JXLH03B strain on pigs, nine 80-day-old healthy pigs were challenged by tracheal injection at doses of 1.2 × 109 , 1.2 × 1010, and 1.2 × 1011 CFU per pig. However, no animals showed clinical symptoms post-challenge, and no abnormalities were observed in the histopathological examination of the pig brain samples (data not shown). Therefore, we failed to establish the infection model of JXLH03B O. anthropi on healthy pigs. In humans, O. anthropi mainly infects immunocompromised patients [1,3,4]. We suspect that the O. anthropi-infected pig could be immunity compromised due to other pathogen infections. Porcine Reproductive and Respiratory Syndrome Virus (PRRSV) was detected in the diseased pig (data not shown). PRRSV is known to cause immunosuppressive respiratory disease [28]. Therefore, we suspect that the pig was infected with O. anthropi due to a compromised immune status. O. anthropi is widely distributed in soil and water [2], so we suspect that the pig could have been infected through these pathways.
O. anthropi is resistant to practically all β-lactams except carbapenems, partly due to the production of an AmpC b-lactamase [29]. Our results showed that the JXLH03B isolate was resistant to penicillin and ceftiofur, which was consistent with the literature reports [30].
《5. Conclusion》
5. Conclusion
In this study, we were the first to isolate and characterize O. anthropi from a pig. This bacteria was able to experimentally infect mice and led to typical meningitis, which indicated that O. anthropi may have a broader host spectrum of infection.
《Acknowledgements》
Acknowledgements
This work is supported by Ten-thousand Talents Program (to Xiangdong Li) and Luoyang Heluo Talent Plan (to Kegong Tian).
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Shijiang Gu, Ruiqing Hou, Shengguo Gao, Zhe Sun, Xiangdong Li, Lufeng Zhai, Yunyun Jin, Qiaoyan Zhu, Yonghong Liao, and Kegong Tian declare that they have no conflict of interest or finacial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2019.08.014.














 京公网安备 11010502051620号
京公网安备 11010502051620号




