《1. Introduction》
1. Introduction
Antimicrobial resistance is now recognized as one of the most serious global threats to human and animal health in the 21st century, and the emergence of multidrug-resistant (MDR) pathogens threatens to cause regression to the pre-antibiotic era [1,2]. Consequently, infections caused by highly resistant Gram-positive organisms, such as staphylococci, streptococci, and enterococci, are considered a major public health issue [3]. In particular, the emergence of methicillin-resistant Staphylococcus aureus (MRSA), vancomycin-resistant Enterococcus faecium (VRE), and β-lactamase-resistant Streptococcus pneumonia have made treatment of the infections very challenging.
More than half of the antibiotics in use target the bacterial ribosome; these inhibitors can paralyze protein synthesis by interacting with the 30S small subunit or 50S large subunit [4–6]. Several ribosome-targeting antibiotics (macrolides, pleuromutilins, lincosamides, streptogramins, and oxazolidinones) are viewed as effective alternatives and last-resort drugs for human use (Table S1 in Appendix A) [2,7,8]. Pleuromutilins were discovered as natural agents in 1951; the C-14 derivatives tiamulin (TIA) and valnemulin are widely used in veterinary medicine, and retapamulin (RET) and lefamulin (LEF) are approved for human use against MRSA or β-lactamase-resistant streptococci [9,10]. Structural characterization has revealed that pleuromutilins can bind to the peptidyl-transferase center (PTC) of 23S ribosomal RNA (rRNA), thereby preventing the correct positioning of acceptor or donor substrates at A-sites and P-sites [10–14].
Accumulating evidence suggests that the resistance of Grampositive pathogens to pleuromutilins is increasing; notably, most isolates show cross-resistance to pleuromutilins, lincosamides, and streptogramins A, thereby exhibiting the PLSA phenotype [15–18]. Resistance to pleuromutilins is commonly attributed to the acquisition of endogenous mutations in 23S rRNA or horizontally transmitted resistance determinants, such as the rRNA methyltransferase Cfr or the antibiotic-resistance adenosine triphosphate (ATP)-binding cassette (ABC)-F family proteins— Vga/Lsa/Sal/VmlR [4,7,19]. Antibiotic-resistance ABC-F family genes have been found in the genomes of numerous pathogens and confer resistance to a broad range of clinically relevant antibiotics targeting the ribosomes [20–22]. Unlike other ABC superfamily members that actively pump drugs out of the membranes, the (ABC)-F family proteins do not fuse to transmembrane domains (TMDs). Accordingly, the mechanism by which ABC-F members mediate antibiotic resistance has been a subject of long-standing controversy. Cryo-electron microscopy (cryo-EM) structures of two ABC-F proteins, MsrE and VmlR, which are bound to the ribosome, indicating that the protection is conferred by interactions with the antibiotic-stalled ribosomes and displacement of the bound drug [23–25].
Streptococcus suis (S. suis) is an emerging zoonotic pathogen; the largest outbreak in humans occurred in China in 2005 (204 infections and 38 deaths) [26]. S. suis may also act as a resistance gene reservoir, contributing to the spread of resistance genes to major streptococcal pathogens [27,28]. During a surveillance study of S. suis resistance in China, 42 TIA-resistant isolates did not harbor known resistance determinants. In this study, we identified and characterized a novel antibiotic-resistance determinant, SrpA (S. suis ribosome protective ABC-F family protein), including analyses of its physicochemical properties and molecular mechanisms. Our comprehensive analyses, which include functional cloning, homology modeling, molecular docking, mutagenic analyses, intracellular antibiotics accumulation assays, and ribosome binding assays, provide a detailed understanding of the mechanisms by which SrpA confers antibiotic resistance.
《2. Materials and methods》
2. Materials and methods
《2.1. Bacterial strains》
2.1. Bacterial strains
A total of 166 S. suis isolates were collected from five provinces in China in 2018, and 72 isolates showed TIA resistance. Escherichia coli (E. coli) BL21, DH5a, S. aureus RN4220, and S. suis SD1BY15 were used as hosts for cloning.
《2.2. Bioinformatics and sequence analysis》
2.2. Bioinformatics and sequence analysis
The genomic DNA of all tested isolates was extracted as described previously [29] and subjected to whole-genome sequencing. A pan-genome analysis was conducted by means of the rapid Roary pipeline for 42 resistant candidates (without known determinants) and 94 susceptible strains [30]. The data were processed using the Python package pandas, and open reading frames (ORFs) annotated as putative ABC transporter ATPbinding proteins in more than ten isolates were extracted. The results were visualized using Matplotlib and Seaborn. MEGA7.0 was used to generate a phylogenetic tree of ABC-F family proteins by the neighbor-joining method (bootstrap: 1000 times) [31]. Sequence logos of corresponding proteins were generated using online tools [32]. The nucleotide sequence of srpA and flanking regions has been deposited in the GenBank database under accession number MT550884.
《2.3. Functional cloning of srpA》
2.3. Functional cloning of srpA
The srpA gene, including 257 base pairs (bp) of its upstream region and 89 bp of its downstream region, was cloned from the genomic DNA of HNBY78 (accession number PRJNA616172) and then inserted into the shuttle vector pAM401. The recombinant plasmid was transferred into the recipients by electrotransformation.
《2.4. Homology modeling and molecular docking of SrpA》
2.4. Homology modeling and molecular docking of SrpA
The three-dimensional (3D) model of SrpA was constructed by means of homology modeling using the SWISS-MODEL web server↑ . SrpA exhibited the highest sequence identity (30%) to the crystal structure of the Thermus thermophilus ribosome-MsrE complex (Protein Data Bank (PDB) ID: 5Zlu.1.u). Based on the GMQE (global model quality estimation) and QMEAN (qualitative model energy analysis) score, model 1 was selected as the best model [33]. The S. aureus ribosome structure was downloaded from the PDB database (5TCU), while the molecular structure of valnemulin was extracted from ChemSpider . Smina was used as a molecular docking tool to investigate the binding among SrpA, valnemulin, and ribosomes. A total of 2000 docking conformations were extracted by a cluster analysis. A small cluster radius of 10.0 Å for the RMSD cutoff, and a smaller interface cutoff of 10.0 Å, generated better clustering results. The top ten poses in the largest clusters were obtained and the results were analyzed using PyMol.
. Smina was used as a molecular docking tool to investigate the binding among SrpA, valnemulin, and ribosomes. A total of 2000 docking conformations were extracted by a cluster analysis. A small cluster radius of 10.0 Å for the RMSD cutoff, and a smaller interface cutoff of 10.0 Å, generated better clustering results. The top ten poses in the largest clusters were obtained and the results were analyzed using PyMol.
↑ http://www.swissmodel.expasy.org
 http://www.ChemSpider.com
http://www.ChemSpider.com
《2.5. Mutational analysis of the extended loop region and ATP hydrolysis glutamates》
2.5. Mutational analysis of the extended loop region and ATP hydrolysis glutamates
SrpA mutants were constructed by means of site-directed mutagenesis using the primers listed in Table S2 in Appendix A. A 6×His tag at the N-terminal end of SrpA was introduced in all primers. Sequencing of the complete srpA gene from each mutant was conducted to ensure that no extraneous mutations occurred. The expression of SrpA in different derivatives was assessed with Western blotting using an anti-His HRP Ab at a dilution of 1:2000.
《2.6. Intracellular antibiotics accumulation assays》
2.6. Intracellular antibiotics accumulation assays
Intracellular accumulation assays were performed, as described previously [34]. S. aureus RN4220, RN4220-pAM401, or RN4220- pAM401-srpA were cultured overnight and 650 μL of each cell solution was transferred into a 1.5 mL Eppendorf tube. After equilibrating at 37 °C with shaking for 5 min, valnemulin at final concentrations of 2 or 16 μg·mL-1 was added. Samples were incubated at 37 °C with shaking for 30 min and were then processed in liquid nitrogen (3 min) followed by 65 °C water (3 min) for lysis. Lysates were centrifuged at 20 000 g for 5 min; the supernatant was diluted 100-fold with water-methanol (2:1, v/v) and vortexmixed prior to liquid chromatography-tandem mass spectrometry (LC-MS/MS) analysis. The source conditions of the MS/MS system were as follows: capillary voltage, 2.5 kV; source temperature, 150 °C; desolvation temperature, 500 °C; cone gas (N2) flow rate, 50 L·h-1 ; and desolvation gas (N2) flow rate, 800 L·h-1 . Valnemulin was analyzed in positive electrospray ionization (ESI) mode. The optimized MS/MS transitions as well as the specific cone voltages and collision energies were as follows: valnemulin, cone voltage 22 V, m/z 263.0 > 163.9 (collision energy, 22 eV). Auto dwell time was applied to ensure that approximately 15 data points were acquired for each chromatographic peak.
《2.7. Ribosome binding assays》
2.7. Ribosome binding assays
The expression and purification of native srpA and its mutants were performed as described previously [29]; in brief, srpA was ligated into pET28a and transformed to E. coli BL21 (DE3) cells. E. coli BL21-containing srpA were cultured overnight, followed by large-scale induction by isopropyl-β-D-thiogalactoside (IPTG) [23]. Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was used to assess the expression of srpA. Proteins were then purified on Ni2+-nitrilotriacetic acid (NTA) affinity resin following a modified version of the manufacturer’s instructions (QIAGEN, Germany) [25]. Ribosomes from S. aureus RN4220 were purified by means of differential centrifugation, as described by Wu et al. [35]. The cells were resuspended in 4 mL of buffer A (10 mmol·L-1 Tris-HCl containing 4 mmol·L-1 MgCl2, 100 mmol·L-1 KCl, and 10 mmol·L-1 NH4Cl, pH 7.2). Cell debris was discarded by centrifugation at 30 000 ×g for 15 min at 4 °C. The supernatant was centrifuged at 100 000 ×g for 120 min at 4 °C to pellet the ribosomes [25].
The fluorescent conjugates (tracer) of valnemulin (VAL) and enrofloxacin (ENR) were prepared as described previously [36]. The concentrations of SrpA and ribosomes were optimized for fluorescence polarization immunoassay (FPIA), and the results are depicted in Fig. S1 in Appendix A. Concentrations of 1000 nmol·L-1 ribosomes and 5 μmol·L-1 SrpA were chosen for further analyses. The antibiotic–ribosome binding assay was performed by means of the addition of 70 μL of VAL-(4,6-dichlorotriazinyl) aminofluorescein (VAL-DTAF) or ENR-aminofluorescein (ENR-AF) to purified ribosomes (1000 nmol·L-1 ) in 70 μL of buffer A (10 mmol·L-1 Tris(pH 7.5), 60 mmol·L-1 KCl, 10 mmol·L-1 NH4Cl, 300 mmol·L-1 NaCl, 6 mmol·L-1 MgCl2, 0.1 mmol·L-1 ATP) at 37 °C for 30 min. Reaction mixtures were then shaken for 10 s in the microplate reader, and the fluorescence polarization (FP) values for the complex were measured at excitation wavelength  = 485 nm, emission wave-ength
= 485 nm, emission wave-ength  = 530 nm, cutoff = 515 nm, and G factor = 1.0, while the blank control only contained buffer A, with the same assay conditions [37]. The ability of SrpA to prevent the binding of VALtracer to the ribosome was assessed as follows. The ribosomes were preincubated in 70 μL of reaction mixture with 5 μmol·L-1 native SrpA, SrpA mutants, or bovine serum albumin (BSA) at 37 °C for 30 min before the addition of VAL-tracers. In competitive experiments, the ribosomes were pretreated with VAL-tracers for 30 min, and then incubated with native SrpA.
= 530 nm, cutoff = 515 nm, and G factor = 1.0, while the blank control only contained buffer A, with the same assay conditions [37]. The ability of SrpA to prevent the binding of VALtracer to the ribosome was assessed as follows. The ribosomes were preincubated in 70 μL of reaction mixture with 5 μmol·L-1 native SrpA, SrpA mutants, or bovine serum albumin (BSA) at 37 °C for 30 min before the addition of VAL-tracers. In competitive experiments, the ribosomes were pretreated with VAL-tracers for 30 min, and then incubated with native SrpA.
《2.8. Determination of the genomic locations of SrpA》
2.8. Determination of the genomic locations of SrpA
S1 nuclease pulsed-field gel electrophoresis (S1-PFGE) and Southern blotting were performed to determine the location of srpA in S. suis. The gel plug of srpA-positive isolates was prepared as described above and subjected to electrophoretic migration for 20 h [38].
The gel was inverted and placed on the gel box, and a nylon membrane, three dry filter papers, and six inches of cut paper towels were placed on the top of the gel for 16 h. The srpA probe was obtained by polymerase chain reaction (PCR) amplification from the genomic DNA of S. suis HNBY78, and then labeled using the digoxin (DIG) high prime DNA labeling and detection starter kit I (Roche, Switzerland).
《3. Results》
3. Results
《3.1. Identification and characterization of a novel pleuromutilin resistance determinant in S. suis》
3.1. Identification and characterization of a novel pleuromutilin resistance determinant in S. suis
During routine surveillance of S. suis resistance profiles in China (2017–2018), 166 isolates were obtained from five provinces, including 94 TIA-susceptible isolates and 72 resistant isolates (64–128 μg·mL-1 ). Interestingly, whole-genome sequencing analysis suggested the presence of the PLSA resistance gene, lsa(E), in 30 TIA-resistant isolates. However, no known resistance determinants were detected in the other 42 strains. Hence, these strains were chosen for an in-depth analysis of the molecular basis of pleuromutilin resistance. A pan-genome analysis was used to compare gene distributions between TIA-susceptible and TIA-resistant isolates, with a focus on ORFs annotated as ATP-binding proteins, which have been implicated in pleuromutilin resistance [20]. As depicted in heatmaps (Fig. 1), 68 putative ATP-binding proteins were observed in all annotated isolates. Most ORFs showed no significant correlation with pleuromutilin resistance. However, an ORF annotated as ‘‘expZ” was only detected in resistant isolates. We then manually inspected the ‘‘expZ” ORFs using the rapid annotation using subsystem technology (RAST)-server and confirmed a 1368-bp ORF in all 42 resistant isolates. A sequence alignment demonstrated that this protein exhibited approximately 36% amino acid sequence identity with another antibiotic-resistance ABC-F protein, Vga(E), and about 31.6% with VmlR (expZ) [39]. To confirm the role of this putative resistance gene, a 1980-bp DNA fragment including this ORF and its flanking region was ligated into the shuttle vector pAM401, and the recombinant plasmid was electroporated into S. suis SD 1B15 and S. aureus RN4220. In comparison with the recipient carrying an empty plasmid, transformants carrying the putative determinant showed significantly higher minimum inhibitory concentrations (MIC) of VAL, TIA, retapamulin (RET), LEF, virginiamycin M1 (VGM), and lincomycin (LIN) (Table 1), while no differences in MIC were detected for vancomycin, erythromycin, kanamycin, gentamicin, tetracycline, linezolid, and florfenicol (FFC). These results suggested that the novel 1368-bp ORF is a new resistance determinant able to mediate cross-resistance to pleuromutilins, lincosamides, and streptogramin A. Accordingly, we named this ORF SrpA (S. suis ribosome protective ABC-F family protein; accession number MT550884).
《Fig. 1》
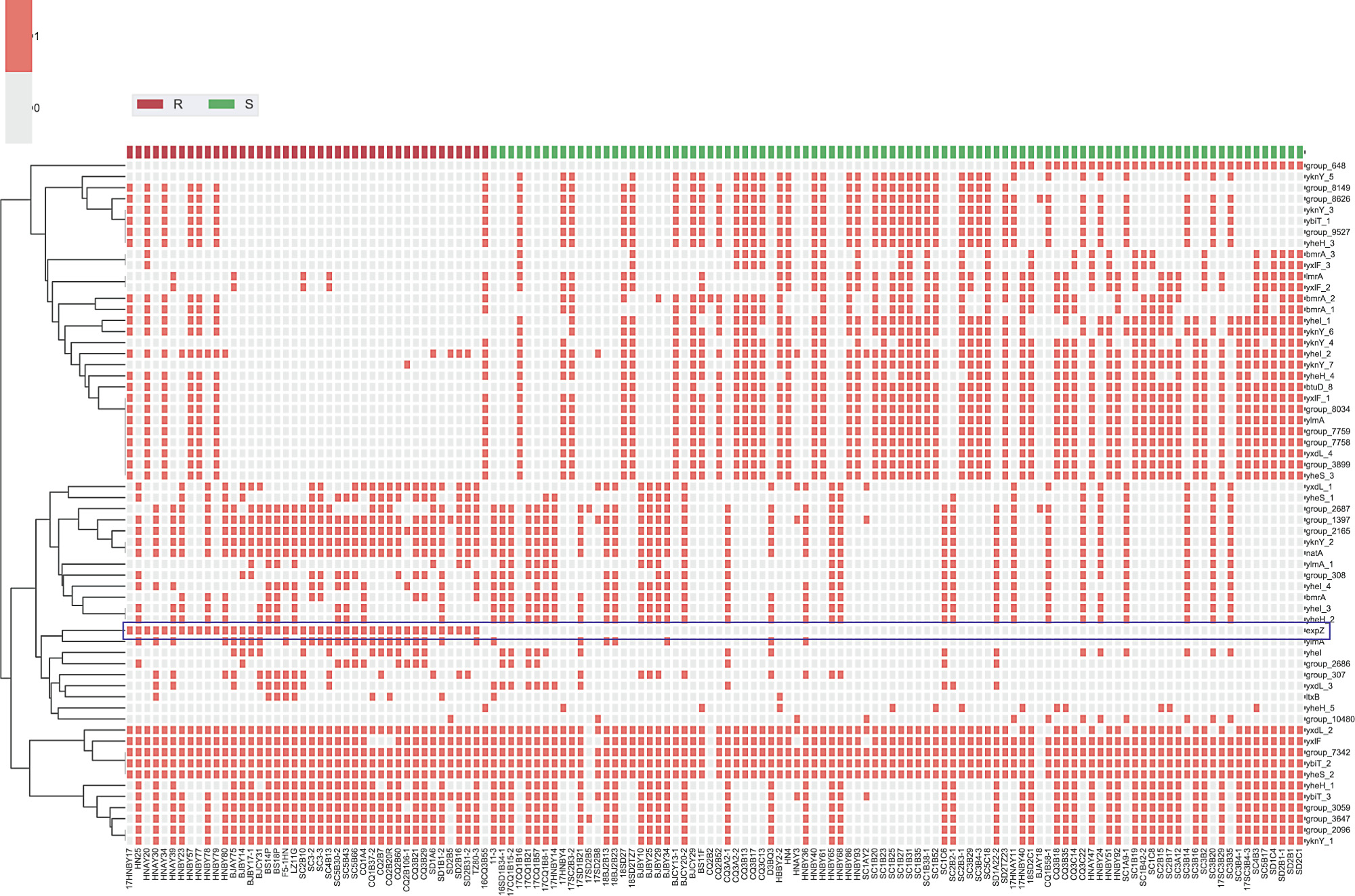
Fig. 1. Distribution of putative ATP-binding proteins in TIA-resistant and TIA-susceptible isolates. Putative ATP-binding proteins are shown along the ordinate. The names of 136 S. suis included in this work are shown on the abscissa. TIA-resistant (R) isolates are shown in red, while susceptible isolates (S) are marked in green. The presence or absence of ATP-binding proteins is indicated by filled or empty squares, respectively. The row ‘‘expZ” is marked with a rectangular border and an asterisk.
《Table 1》
Table 1 Minimum inhibitory concentrations of tested antimicrobial agents for the studied bacterial isolates.
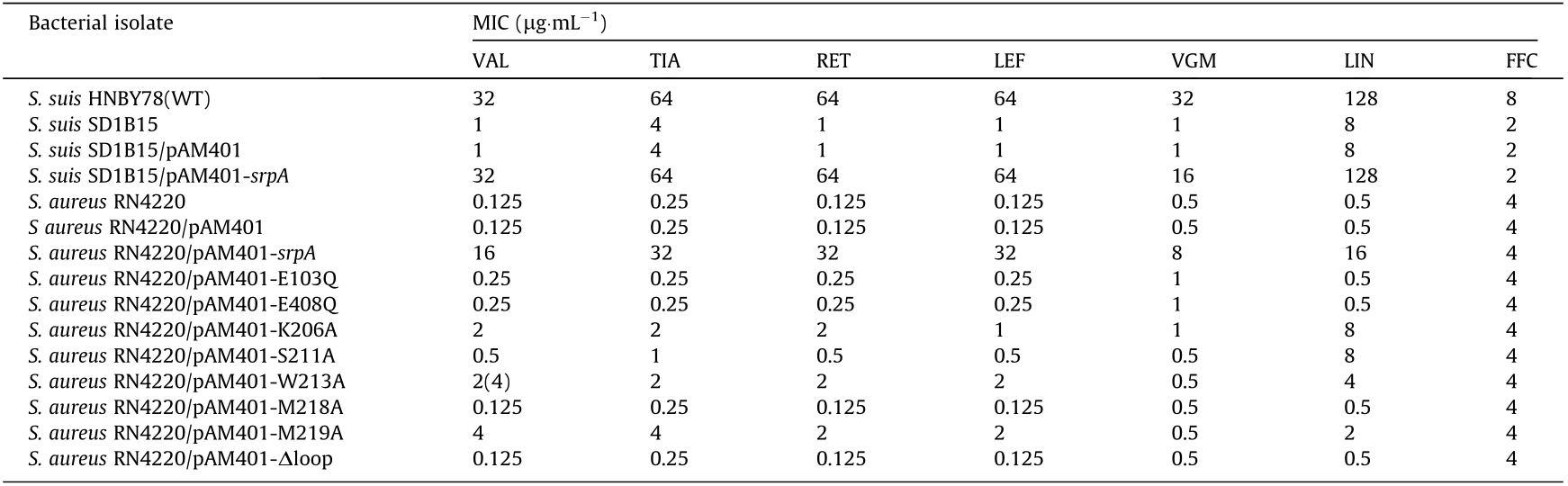
VGM: virginiamycin M1; WT: wild type.
《3.2. Relatedness of SrpA to other antibiotic-resistance ABC-F proteins》
3.2. Relatedness of SrpA to other antibiotic-resistance ABC-F proteins
A SMART↑ sequence analysis predicted that SrpA has the canonical architecture of ABC-F family proteins, with two Walker A motifs (residues 36–44 and 303–311), two Walker B motifs (residues 98– 102 and 403–407), two ABC signatures (residues 78–83 and 383– 388), two H-loop switches (residues 132–437), and a lack of a transmembrane-associated domain (Fig. S2 in Appendix A). Furthermore, for the catalytic motifs of SrpA, the consensus signature sequence was LSGGE, instead of the LSGGQ in most ABC superfamily proteins [40].
↑ http://smart.embl-heidelberg.de/
Thereafter, we built a neighbor-joining tree to estimate the relationships of SrpA with other antibiotic-resistance ABC-F family proteins. As depicted in Fig. 2, the phylogenetic tree was characterized by five deep-branching lineages. SrpA belonged to a lineage that includes Vga homologs, Msr homologs, VmlR, and Lmo0919. A sequence alignment showed that SrpA shares over 30% amino acid identity with the loci in this lineage, and the closest homolog was Vga(E) in S. aureus (36%). Although antibiotic-resistance ABC-F family proteins are found in a variety of bacterial groups, SrpA is the first antibiotic-resistance ABC-F protein found in S. suis.
《Fig. 2》
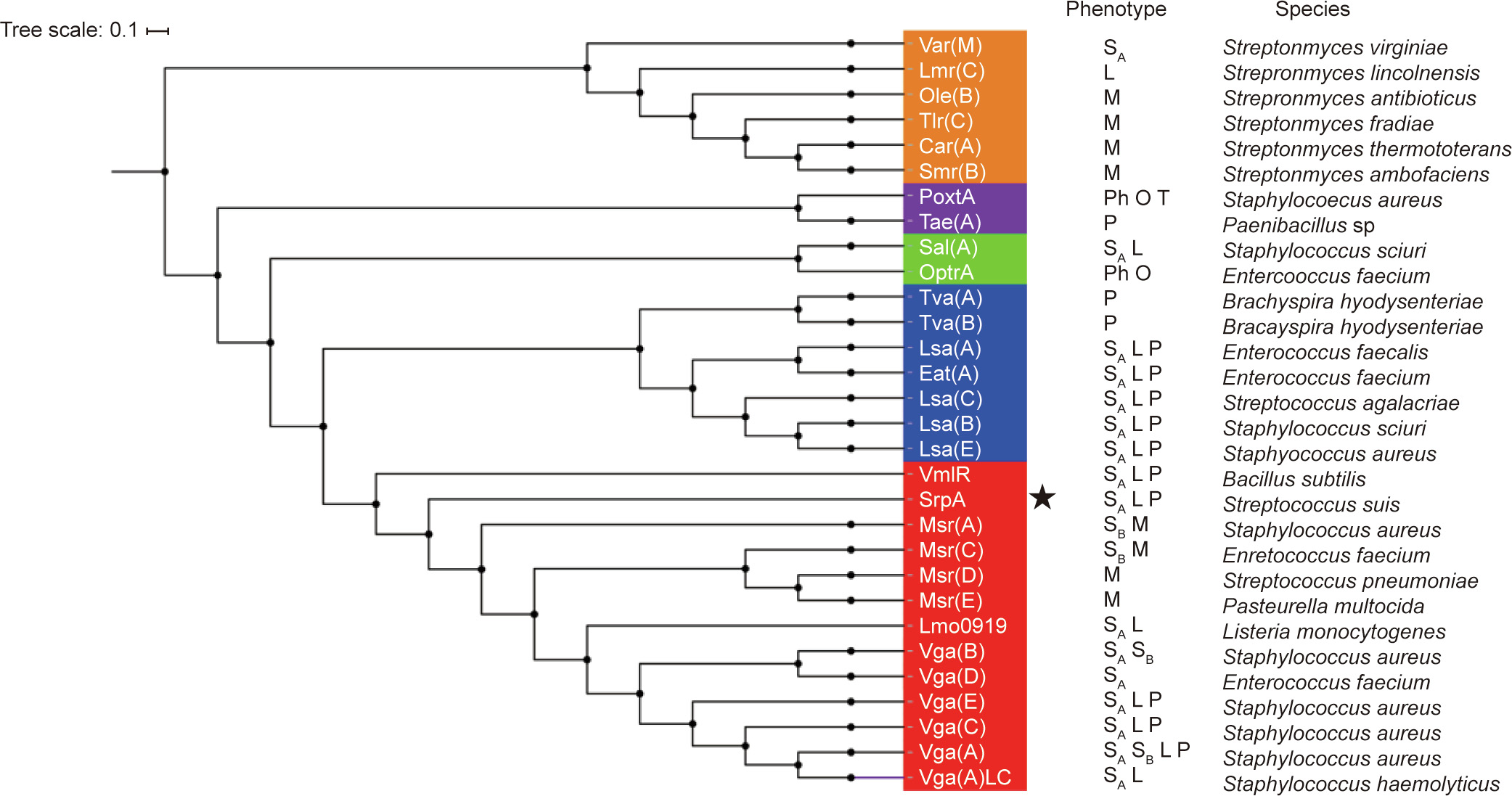
Fig. 2. Phylogenetic tree of antibiotic-resistance ABC-F proteins constructed using the neighbor-joining method. The tree represents the consensus obtained after 1000 replicates. Amino acid sequences were extracted from National Center for Biotechnology Information (NCBI). Antibiotics affected by the different proteins are indicated, and the species in which the various antibiotic-resistance ABC-F have been described for the first time are also indicated. SA: streptogramin A; SB: streptogramin B; P: pleuromutilins; L: lincosamides; M: macrolides; Ph: phenicols; O: oxazolidinones; T: tetracyclines; the novel antibiotic-resistance ABC-F determinant SrpA is indicated by an asterisk.
《3.3. Homology modeling and molecular docking of SrpA》
3.3. Homology modeling and molecular docking of SrpA
A 3D model of SrpA was created using the SWISS-MODEL tool, and the crystal structure of Msr(E) (PDB ID: 5Zlu.1.u), which shared 30% sequence identity with SrpA, was selected as the most suitable template. The structure of SrpA was predicted to possess conserved features of antibiotic-resistance ABC-F family proteins, such as two ABC transporter domains (ABC1 and ABC2) carrying highly conserved nucleotide-binding sites (NBSs; residues 36–43 and 303– 310) assembled at its base, and a domain linker (residues 163– 250) that forms two long crossed helices (α1 and α2) connected by an extended loop (residues 203–224; see Fig. 3(a)) [20,25].
《Fig. 3》
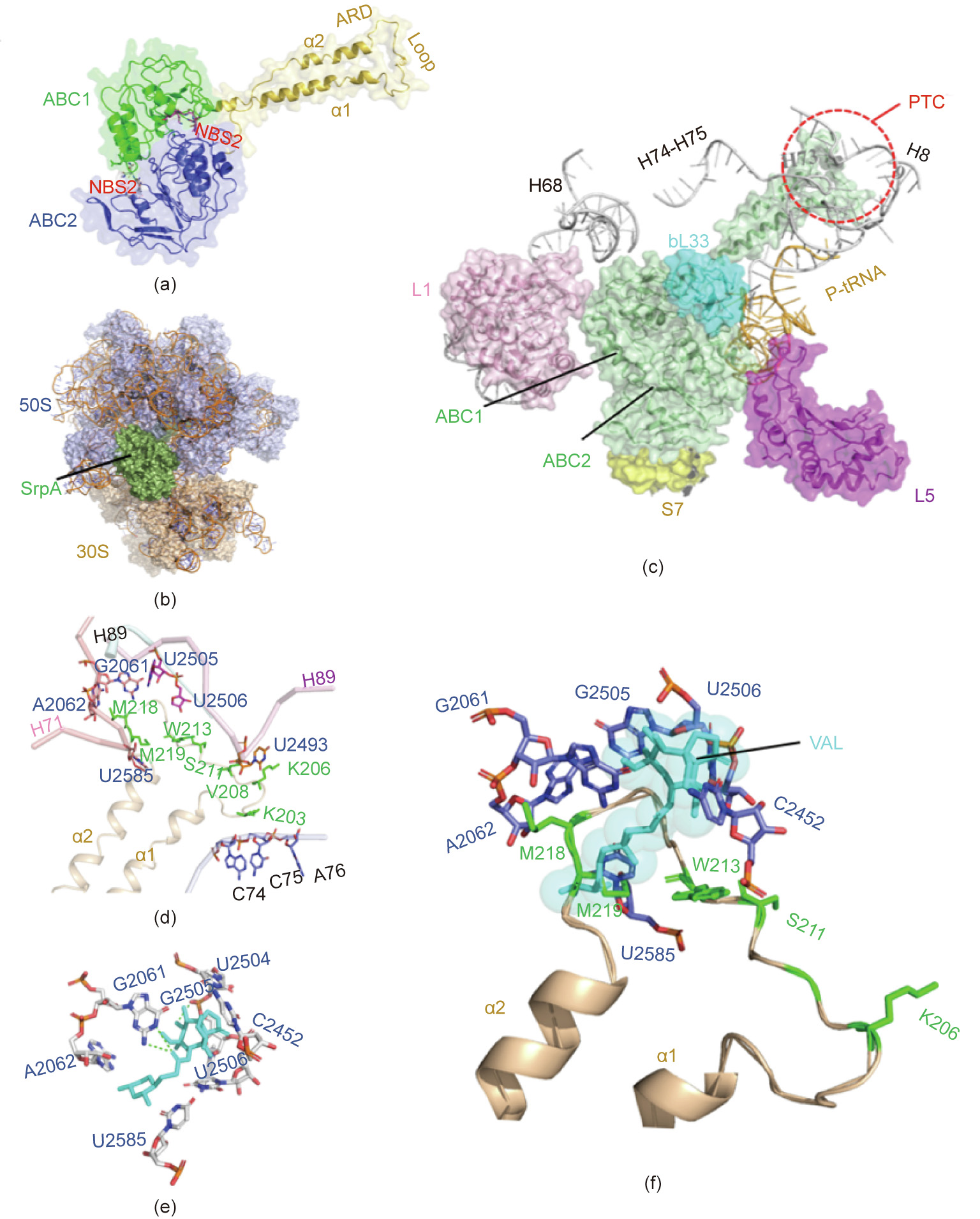
Fig. 3. Homology modeling and molecular docking of SrpA. (a) 3D structures of SrpA generated by the SWISS-MODEL server; protein shown in cartoon with secondary structure elements of ABC1 (green), ABC2 (blue), and domain linker (yellow orange) labeled. (b) Binding mode of SrpA (green), and ribosomes (50S slate, 30S light orange). (c) Interactions between the SrpA (green) ABC1 domain and the 50S subunit r-protein L1 (pink), bL33 (cyan), 23S rRNA helices H68, H73–H75, and H89 (white), and between the ABC2 domain and the 30S subunit r-protein S7 (yellow), r-protein L5 (purple), and tRNA (wheat). (d) Orientation of the SrpA extended loop (yellow orange) and surrounding key PTC residues. (e) TIA-binding pocket (cyan) in the TIA-ribosome complex. (f) Superposition of ribosome-bound TIA onto the SrpA-ribosome complex.
Several studies have demonstrated that antibiotic-resistance ABC-F proteins bind to the ribosome and allosterically dissociate the drug from its binding site [23,24]. In the present work, molecular docking was used to analyze SrpA-ribosome interactions. The crystal structures of the S. aureus 70S ribosome were extracted from the PDB database (ID: 5TCU), and the complex was docked with a 6° Euler angle sampling size using ZDOCK 3.0.2 (Fig. 3(b)). As shown in Fig. 3(c), the ABC1 domain of SrpA faces the L1 stalk of the 50S subunit, whereas the ABC2 domain contacts the r-protein L5, S7, and the elbow of peptidyl-tRNA(P-tRNA). In contrast to the few ribosome contacts observed for the two ABC domains, the elongated domain linker established extensive contacts with ribosomes, as it stretched in parallel with H74–H75 of 23S rRNA and the acceptor arm of P-site tRNA. In addition, the tip loop region was inserted deep into the PTC, approaching the 23S rRNA helices H89 and H73. The residues Lys203, Arg204, and Lys205 in the loop region directly contacted the CCA end of a P-site tRNA, while the Lys206–Ser224 residues and their side chains interacted with the flexible nucleotides U2585, A2062, G2061, U2505, U2506, and U2493 (Fig. 3(d)). Interestingly, the aforementioned nucleotides seemed to constitute the VAL-binding pocket, as the VALribosome docked compounds demonstrated that G2061 and U2505 can hold the two sides of VAL by hydrogen bonds, while the other residues (e.g., U2585, A2062, U2506, and C2452) were also located in close proximity to VAL (Fig. 3(e)). Superimposing the SrpA-ribosome complex and the VAL-ribosome complex revealed steric overlap between the SrpA loop region and VAL in the VAL-binding pocket; in particular, the Met219 residue directly clashed with the C14 extensions of VAL (Fig. 3(f)).
《3.4. Mutagenic analyses of the ATP hydrolysis site and extended loop region》
3.4. Mutagenic analyses of the ATP hydrolysis site and extended loop region
ATP hydrolysis appears to play a key role in the functioning of ABC-F proteins, as ATP hydrolysis-deficient energy-dependent translational throttle A (EttA) could not be released from ribosomes, and ATP hydrolysis-deficient MsrE and Vga(A) show reduced or abolished antibiotic resistance [23,40,41]. In this work, the conserved catalytic glutamates following the Walker B motifs in both nucleotide-binding domains (NBDs) (Glu103 in NBD1 and Glu in 408 NBD2) were mutated to glutamine. A growth curve showed that this ATP hydrolysis-deficient SrpA mutant (E103Q/ E408Q) did not significantly affect the cell growth of S. aureus (Fig. S3 in Appendix A). However, the antibiotic resistance of the SrpA 103Q/E408Q mutant was significantly lower than that of the wild type, indicating that ATP hydrolysis was crucial for PLSA resistance (Table 1).
The extended loop regions of antibiotic-resistance ABC-F proteins are collectively referred to as the antibiotic-resistance domain (ARD), as the tip loop region accesses the PTC of the ribosome and directly encroaches on the binding site of PTC-targeting antibiotics [21,42]. Based on previous studies and the insights gain from the docked compounds, the loop region of SrpA appears to play a major role in the efficiency and specificity of antibiotic resistance.
Based on multiple sequence alignment of SrpA, Vga homologs, Msr homologs, VmlR, and Lmo0919 obtained using ClustalX, this short stretch corresponded to residues Lys203–Ser224 in SrpA. Consensus sequence logos were generated for the extended loop region (Fig. 4(a)). A total of four highly conserved amino acid residues were found in SrpA: Lys203, Lys206, Gly207, and Ala217. The arrangement of amino acids at other sites was polymorphic, especially for residues Val208, Trp213, Met218, Gly220, and Ser221. To further confirm the effect of the extended loop region on the efficiency and specificity of antibiotic resistance, we generated mutations in the SrpA loop region at positions Trp206Ala, Ser211Ala, Trp213Ala, Met218Ala, and Met219Ala. The introduction of point mutations at Lys203, Val208, Gly220, and Ser221 was not successful, as the mutants were quite unstable. In addition, recombinant vector pAM401 with the truncation of the extended loop region (residues Lys203–Ser224, △loop) was constructed by homologous recombination.
《Fig. 4》
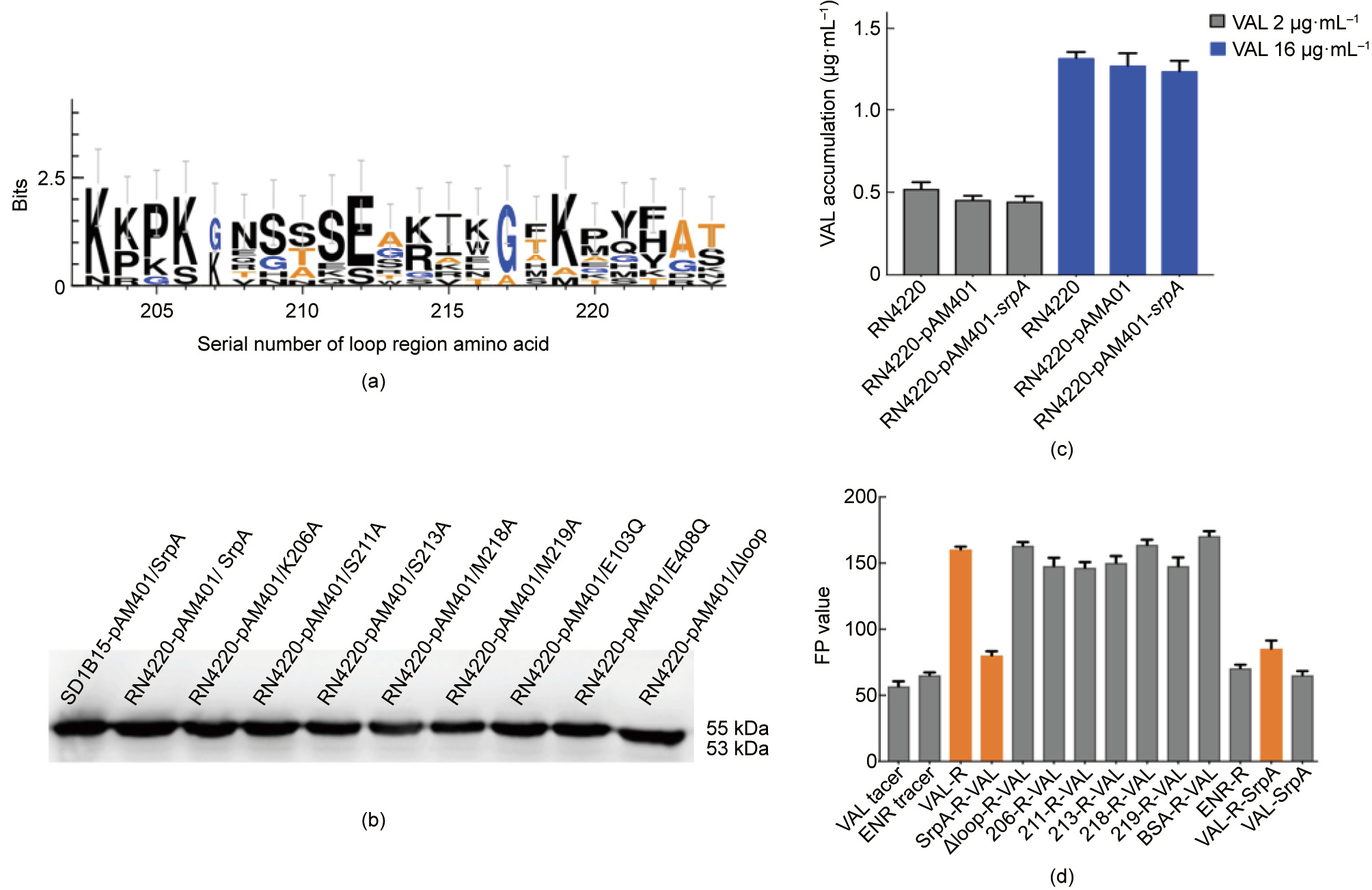
Fig. 4. Key amino acids of SrpA for ribosome protective activity. (a) Sequence logos of the loop region amino acid residues generated for SrpA, Vga(A), Vga(E), Msr(E), VmlR, and Lmo0919. The height of each letter is proportional to the frequency of the corresponding amino acid residues (the serial number of amino acid as indicated in the x-axis) in a given site. (b) Western blotting analysis of native SrpA and SrpA mutants in S. aureus RN4220. (c) Intracellular antibiotics accumulation assay (samples treated with 2 μg·mL-1 VAL are indicated in grey; those treated with 16 μg·mL-1 VAL are indicated in blue). (d) Ribosome binding assay: columns 1 and 2 show the FP values of VAL and ENR tracer; in column 3, 1000 nmol·L-1 purified S. aureus ribosomes were added to the system; in column 4, the ribosomes were preincubated with 0.5 μmol·L-1 SrpA for 30 min; in columns 5–11, SrpA was replaced with mutants (K206A, S211A, W213A, M218A, M219A, and △ (K203–S224)), or BSA; in column 12, 1000 nmol·L-1 of purified S. aureus ribosomes were added to the ENR-tracer system; in column 13, the purified S. aureus ribosomes were first pretreated with VAL-tracer for 30 min and 5 μmol·L-1 SrpA was introduced to the complex; in column 14, 5 μmol·L-1 SrpA was added directly to the VAL-tracer without ribosomes. The results are the means of three independent repeats; error bars represent standard deviation (SDs). No ribosome addition was included as a control. R: S. aureus ribosomes.
An antibiotic susceptibility test showed that mutations in residues in the loop domain did indeed influence the PLSA resistance phenotype in vivo (Table 1). Compared with isolates harboring intact SrpA, strains bearing mutations at residues 206, 211, 213, 218, and 219 showed a two- or four-fold decrease in LIN resistance and an eight- to 16-fold decrease in the MIC of virginiamycin. For pleuromutilin compounds, strains carrying the Trp206Ala, Trp213Ala, or Met219Ala substitutions exhibited two- to 16-fold decreases in the MICs of VAL, TIA, RET, and LEF. The Ser211Ala substitution resulted in 16- to 32-fold lower MICs. Intriguingly, the truncation of the extended loop region (residues Lys203–Ser224, Dloop) or even the mutation of a single residue (Met218Ala) could completely abolish the ability to mediate PLSA resistance. As a control, the FFC resistance of all derivates remained unchanged. Taken together, these results suggest that the extended loop region, particularly Met218, is crucial for PLSA resistance in vivo.
A Western blotting analysis with an anti-6×His antibody was used to examine the expression of SrpA derivatives in the transformants. A single protein band with a molecular weight of about 55 kDa was detected in the positive control S. aureus RN4220 harboring native SrpA and its seven mutant derivatives (Trp206Ala, Ser211Ala, Trp213Ala, Met218Ala, Met219Ala, Glu103Gln, and Glu408Gln). The transformant carrying the srpA-△loop migrated shows a protein size of 53 kDa (Fig. 4(b)); this size decrease might be explained by the deletion of the extended loop region.
《3.5. Intracellular antibiotics accumulation assays and ribosome binding assays》
3.5. Intracellular antibiotics accumulation assays and ribosome binding assays
To further elucidate the action mechanism of SrpA, an intracellular antibiotics accumulation approach and a modified ribosome binding assay were used in this work.
The results of the time-course study of VAL acumulation are shown in Fig. 4(c). Representative ultra high performance liquid chromatography (UHPLC)-MS/MS chromatograms of VAL in tested isolates are shown in Fig. S4 in Appendix A. The original recipient strain S. aureus RN4220 and the two transformants carrying the empty pAM401 plasmid or pAM401/srpA exhibited similar trends in VAL accumulation (add 2 and 16 μg·mL-1 VAL, respectively), which indicated that srpA might not act as an antibiotic transporter.
Since more evidence suggests that the mediated resistance of these ARE ABC-F proteins could occur through a ribosome protection mechanism, a modified FPIA was used to measure the interaction between SrpA and ribosomes. The FP value is determined by the size of the fluorescent-labeled antibiotics tracer. For an unbound tracer with a small size and fast Brownian rotation, the FP value is low, whereas for the tracer–ribosome complex, the FP value is high. The initial FP values of the tracers VAL-DTAF and ENR-AF were 58 and 66, respectively (Fig. 4(d)). After the introduction of 1000 nmol·L-1 of purified S. aureus ribosomes, the FP value of the Val-ribosome mixture rose to 160. For the ENR mixture, the FP value showed nearly no variation, suggesting specific binding between VAL and bacterial ribosomes.
Interestingly, when S. aureus ribosomes were preincubated with SrpA (5 μmol·L-1 ) for 30 min and subsequently titrated into VALDTAF, the FP value of the complex decreased to approximately 79, far less than the value for the direct VAL-ribosome interaction, revealing that SrpA can interact with ribosomes and prevent the subsequent binding of VAL-tracer. When the ribosome was pretreated with VAL-tracer for 30 min and the complex was incubated with SrpA (5 μmol·L-1 ) for another 30 min, the FP value was nearly 88, indicating that SrpA displaces prebound VAL-tracer from ribosomes. When S. aureus ribosomes were pretreated with BSA instead of SrpA, the FP value was similar, confirming that the ability to rescue ribosomes from VAL binding was a specific property of SrpA. The extended loop region plays an important role in mediating antibiotic resistance, and this result was mirrored by FPIA. The preincubation of ribosomes with the SrpA-△loop mutants did not reduce the level of ribosomally associated VAL-tracer. Furthermore, our data demonstrated the importance of the loop residues Trp206, Ser211, Trp213, Met218, and Met219, as mutations to Ala caused a significant loss of ribosomal protection activity.
《3.6. Characterization of srpA in S. suis》
3.6. Characterization of srpA in S. suis
srpA-positive S. suis was collected from five provinces separated by great geographical distance, indicating the wide distribution of this gene in China. S1-PFGE and Southern blotting revealed that srpA was located in the chromosomal DNA of all positive isolates (Fig. S5 in Appendix A). Isolates were subdivided into two types according to genetic context. The type I synteny was identical in 40 isolates, with a 2078-bp penicillin binding protein gene pbp2b located immediately downstream of the srpA gene and an 804-bp mannose-binding lectin (MBL) fold metallo-hydrolase gene yycJ upstream of srpA (Fig. 5). Interestingly, these two motifs were highly conserved in S. suis, even in isolates without srpA, and no transposons or insertion element sequences were identified in the vicinity of srpA. Type II isolates showed two directly repeated ISSsu8-like elements (93% identity) in the region flanking srpA. Reverse PCR confirmed that the insert elements bracketing srpA could recombine and form a 3195-bp circular intermediate including srpA and one copy of the ISSsu8-like locus.
《Fig. 5》
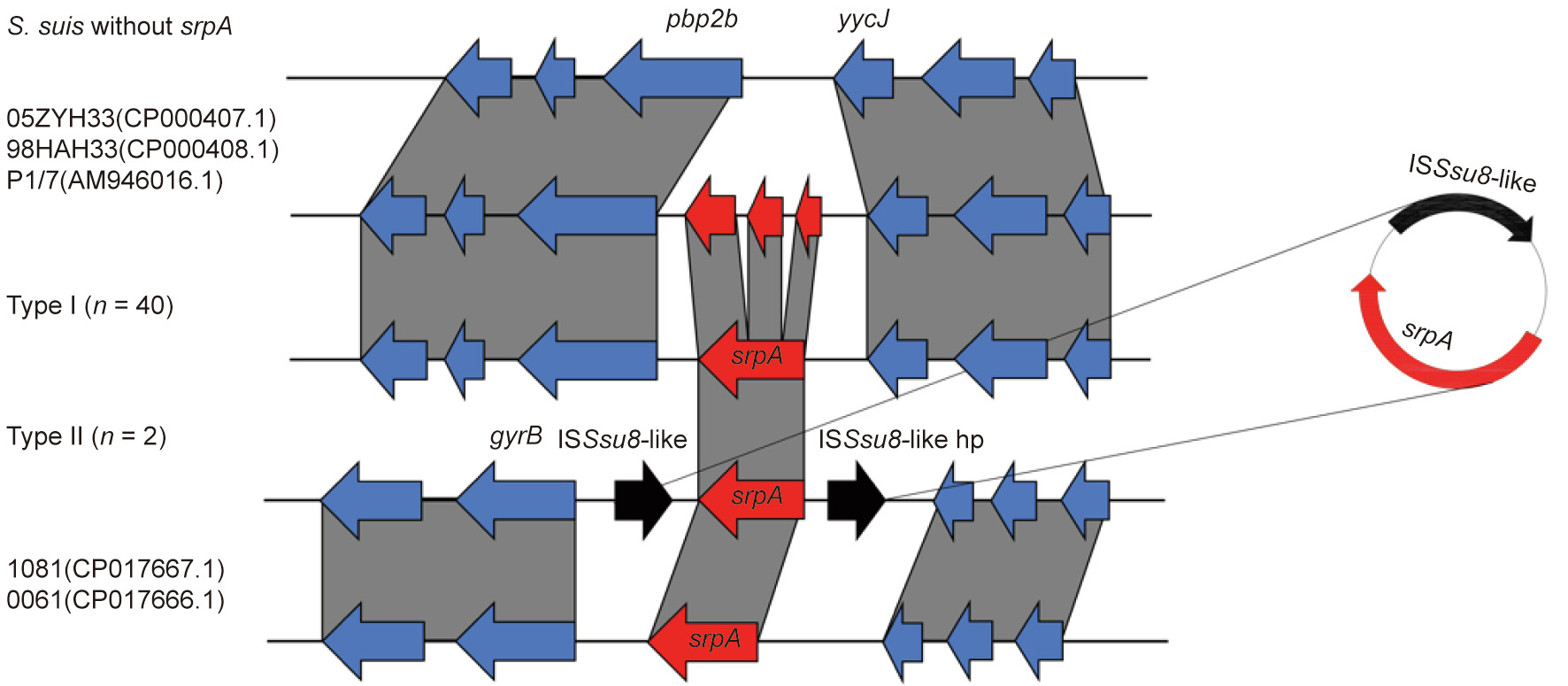
Fig. 5. Genetic context of the srpA gene. Regions with > 90% nucleotide identity are connected by grey zones. srpA is shown in red, insertion sequence (IS) elements in black, and other genes in blue. S. suis 05ZYH33(CP000407.1), 98HAH33(CP000408.1), P1/7(AM946016.1), 1081(CP017667.1), and 0061(CP017666.1) were obtained from NCBI. hp: hypothetical protein.
《4. Discussion》
4. Discussion
Since the discovery of penicillin in 1929, antimicrobial agents have been indispensable for the control of bacterial infections in clinical settings and in veterinary medicine [43]. However, their efficacy has been hampered by the emergence of antimicrobial resistance in bacteria. Many infections are caused by highly resistant Gram-positive organisms, and the emergence of MRSA, VRE, and β-lactamase-resistant streptococci has severely limited the antibiotic arsenal.
To counteract these MDR bacteria, ribosome-targeting antibiotics have been developed as alternative or adjuvant agents in combination with β-lactams or vancomycin [3]. Ribosomes are the protein-synthesizing factories of the cell, and more than half of the antibiotics used in clinical settings target the bacterial ribosome [6]. Despite their diverse chemical structures, all ribosomal active antibiotics interact with the ribosome PTC or the nascent peptide exit tunnel region and possess overlapping binding sites (A-, P-, and E-sites) [4,7,12].
S. suis is an important pathogen in the pig industry and is an agent of zoonosis, transmitted by close contact with infected pigs or pork-derived products [44–46]. In 1998 and 2005, two largescale outbreaks of human S. suis infections occurred in China, with high morbidity and mortality [47]. During a surveillance study of S. suis resistance in China, we found a novel antibiotic-resistance ABC-F resistance determinant named srpA. The ORF encodes a protein of 461 amino acids and confers cross-resistance to pleuromutilins, lincosamides, and streptogramin A in S. suis and S. aureus. Remarkably, all srpA-positive transformants showed high levels of resistance to the newly approved pleuromutilin derivate LEF (2019, Food and Drug Administration (FDA)). LEF is the first pleuromutilin approved for systemic human use and is regarded as a potential treatment option for MRSA or MDR streptococci; accord ingly, LEF resistance in S. aureus and S. suis is an alarming observation.
A sequence alignment of SrpA with other antibiotic-resistance ABC-F family proteins revealed that SrpA belongs to a lineage containing Vga homologs, Msr homologs, Lmo0919, and VmlR. The tertiary structure of SrpA was more similar to that of MsrE, including two ATP-binding domains connected by an 87-amino acid linker. Furthermore, SrpA lacked the C-terminal extension (CTE) found in VmlR (residues 483–547) and Vga homologs (residues 460– 520), potentially explaining the shorter length of SrpA (461 aa) in comparison with these other proteins (521–547 aa) [22].
ATPase activity is important for the function of some ABC-F family members. Mutations of the catalytic glutamate residues following the Walker B motif in EttA influence cell growth by inhibiting protein synthesis, while in Vga(A)LC and Lsa(A), hydrolytically inactive mutants inhibit the peptidyl-transferase activity of the ribosome [22,40]. In the case of MsrE, the glutamates of both LSGGE motifs are positioned close to the 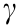 -phosphate of adenylyl imidodiphosphate (AMP-PNP) and are likely involved in ATP hydrolysis, while glutamate mutants show reduced ribosome binding in vitro and azithromycin (AZM) resistance in vivo [23]. In the present work, the SrpA Walker B (E103/408Q) mutants did not show PLSA resistance, suggesting that ATP hydrolysis is pivotal for SrpA activity.
-phosphate of adenylyl imidodiphosphate (AMP-PNP) and are likely involved in ATP hydrolysis, while glutamate mutants show reduced ribosome binding in vitro and azithromycin (AZM) resistance in vivo [23]. In the present work, the SrpA Walker B (E103/408Q) mutants did not show PLSA resistance, suggesting that ATP hydrolysis is pivotal for SrpA activity.
Molecular docking was used to obtain insight into the SrpAribosome interaction. Based on the complex model, ABC1 and ABC2 of SrpA established massive interactions with ribosomal protein and the elbow of the P-tRNA, while the interdomain linker, especially the extended loop region, projected deep into the ribosome PTC, interacted with the CCA arm of P-tRNA, and was located in proximity to the 23S rRNA helices H89 and H73. The architecture and conformation of the SrpA-ribosome interactions were very similar to those of MsrE/VmlR, suggesting that the binding site of antibiotic-resistance ABC-F proteins is highly conserved [23,24]. VAL-ribosome docked compounds revealed that the tricyclic mutilin core on the base of C2452, U2505, U2506, A2062, G2061, and its C14 extension was stabilized by U2585 [2,13,14]. As the SrpA loop residues Met218, Met219, and Trp213 were in direct contact with the crucial nucleotides A2062, G2061, U2585, and C2452 surrounding the VAL molecule, the insertion of the SrpA loop region might cause conformational changes of the VALbinding pocket. Furthermore, Met219 of SrpA directly competed with VAL, similar to Leu242 in MsrE.
The linker connecting the two tandem ABC domains is a defining feature of the ABC-F family, and the loop region of the linker forms stereospecific contact near the PTC in the ribosome [20]. Unlike MsrE and VgaALC variants, truncation of the loop region completely abolished resistance to pleuromutilins, lincosamides, and streptogramin A [22,23]. Furthermore, Lys206Ala, Ser211Ala, Trp213Ala, Met218Ala, and Met219Ala substitutions reduced PLSA resistance to different degrees. This variation is presumably because mutations reduce the size of the sidechain that penetrates most deeply into the PTC in SrpA, thereby eliminating steric clashes with VAL and reducing the magnitude of the relatively small allosteric conformational changes in the PTC upon SrpA binding [20].
The mechanism by which ABC-F proteins mediate antibiotic resistance has been a subject of long-standing controversy. The efflux hypothesis posits that ABC-F proteins export antibiotics out of the cell, similar to other ABC superfamily members, while a recent study has shown that ABC-F proteins mediate antibiotic resistance by interacting with the ribosome and displacing the bound drug [20,25]. In this study, we observed that the presence of srpA had no effect on intracellular antibiotics accumulation. In the follow-up experiment, we also found that the addition of the putative efflux pump inhibitor carbonyl cyanide mchlorophenylhydrazone (CCCP) (10 μg·mL-1 ) or reserpine (20 μg·mL-1 ) had no impact on PLSA resistance in isolates harboring srpA, while for the control strain S. aureus G7 harboring the overexpression fluoroquinolone efflux pump gene NorA, the MIC value of ciprofloxacin decreased from 32 to 4/8 (Table S3 in Appendix A) [48]. Remarkably, a ribosome binding assay unambiguously demonstrated that SrpA can prevent the binding of antibiotics to ribosomes and displacing ribosome-bound antibiotics. In general, all these results indicate that the novel gene srpA does not function as an efflux pump that exports antibiotics out of the cell; rather, it acts like other ABC-F family determinants through a ribosome protection mechanism.
A BLASTp search of the National Center for Biotechnology Information (NCBI) database revealed that SrpA is exclusively found in S. suis, with a number of SrpA-like proteins (82%–99% aa identity) distributed across isolates from China, the United States, the United Kingdom, Vietnam, and the Netherlands, indicating the global dissemination of SrpA. Interestingly, in most isolates, such as 05ZYH33 (CP000407.1), 98HAH33 (CP000408.1), and P1/7 (AM946016.1), srpA could be divided into three adjacent ORFs separated by 100–200 bp, respectively. The first ORF (51 amino acid) showed 88% amino acid identity to SrpA (residues 1–16); the second ORF (180 aa) showed 89% identity (residues 34–212); and the last ORF (249 aa) showed 81% identity (residues 218–455). We speculate that a loop truncation explains the pleuromutilin susceptibility in these strains.
In the present study, all srpA genes were located on chromosomes and were intact. With respect to genetic context, most of the isolates belonged to type Ⅰ (n = 40), in which srpA was invariably located between pbp2b (WP_105203009.1) and yycJ (WP_024376691). However, isolates without srpA harbored a similar arrangement, separated by 400–500 bp. Only some of the antibiotic-resistance ABC-F proteins mediating the PLSA resistance phenotype have been found to be chromosomally encoded and embedded in genes, including vmlR in Bacillus subtilis, lsa(A) in Enterococcus faecalis, tva(A) in Brachyspira hyodysenteriae, and lsa (A) and salA in Staphylococcus sciuri [38,49–51]. The genetic context of type Ⅱ isolates was highly divergent from that of type Ⅰ isolates, with two directly repeated IS481 family ISSsu8-like elements in the flanking region of srpA; this segment could form a circular intermediate composed of srpA and an ISSsu8-like element. Since S. suis has been recognized as a reservoir for the spread of resistance genes to major streptococcal pathogens, the potential risk of the dissemination of srpA from S. suis to other Streptococcus spp. is worrisome [27,52]. The closest NCBI matches to the type Ⅱ segment were regions in S. suis 1081 and S. suis 0061 from China (CP017667.1 and CP017666.1; 71% amino acid identity), but the ISSsu8-like elements were absent from both. In light of these findings, we speculate that srpA might stem from S. suis and confer innate resistance to PLSA. It is conceivable that over evolutionary timescales, some S. suis isolates stably inherited the resistance gene from a common ancestor, while other isolates lost the ancestral srpA gene. Given that S. suis is currently exposed to numerous PLSA antibiotics in veterinary clinics, the appearance of srpA on mobile genetic elements is a feasible survival strategy in PLSA-susceptible S. suis.
In summary, in this study, a new antibiotic-resistance ABC-F family determinant, srpA, was detected; the gene confers crossresistance to pleuromutilins, lincosamides, and streptogramin A in S. suis and S. aureus. SrpA possesses the characteristic ABC-F family protein conformation and shows the highest similarity to Vga (E). Molecular docking suggested that the antibiotic-resistance domain loop region of SrpA penetrates deep into the PTC cavity and occupies the VAL-binding pocket. Mutagenic studies implicated the ARD loop region in mediating the specificity and efficiency of antibiotic resistance. Furthermore, ATP hydrolysis residues (103E/408E) played a paramount role in SrpA activity. Importantly, we found that SrpA can protect ribosomes and promote the dissociation of the drug from its binding sites, suggesting that a similar ribosomal protection mechanism is shared by antibiotic-resistance ABC-F family proteins.
《Acknowledgments》
Acknowledgments
We are grateful for the sampling support from microbiologists in these agencies: Chongqing Academy of Animal Sciences, Sichuan Provincial Agricultural Department, Sichuan Agricultural University, Qingdao Agricultural University, and Henan Agricultural University. The study was supported by grants from the National Key Research and Development Program of China (2016YFD0501304 and 2016YFD0501305). This work was supported by the grants from the National Natural Science Foundation of China (31722057).
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Chaoyang Zhang, Lu Liu, Peng Zhang, Jingpo Cui, Xiaoxia Qin, Lichao Ma, Kun Han, Zhanhui Wang, Shaolin Wang, Shuangyang Ding, and Zhangqi Shen declare that they have no conflict of interest or financial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2020.12.015.














 京公网安备 11010502051620号
京公网安备 11010502051620号




