《1. Introduction》
1. Introduction
Tumor antigens, which are generally categorized as either tumor-associated or tumor-specific antigens, can be recognized by T cells [1]. Upon activation, T cells distinguish tumor cells from normal cells and initiate a cascade of targeted reactions that ultimately eradiate tumoral cells [2]. Previous reports indicate that a majority of tumor antigens arise from the coding regions of the genome, as exemplified by tumor-specific single-nucleotide variants (SNVs) [3,4], insertions and deletions (collectively called InDels) [5], and gene fusions [6]. However, in validation experiments, only a minority of the tested peptides (around 2%) have been found to be immunogenic in different cancer types [3,4].
Circular RNAs (circRNAs) are a type of mainly stable endogenous RNAs with a covalently closed continuous loop that is generated via backsplicing, in which the 30 splice site links to the upstream of the 50 splice site [7,8]. Previous in vitro and in vivo studies have demonstrated the existence of translated circRNAs and tumor-specific circRNAs [7,9,10]. In circRNAs, highly conserved open reading frames (ORFs) encode peptides in a way that is independent of the 50 cap structure, such as internal ribosome entry site (IRES) induction [11,12]. Moreover, the functional peptides encoded by novel ORFs at the backspliced junctions most likely mismatch with normal protein-coding transcripts, so the candidate antigen peptides derived from circRNAs have a high likelihood of being recognized as foreign by T cell receptors (TCRs).
The number of candidate antigen peptides is overwhelming for clinical trials, and these candidates are rarely shared among patients [3,4,13]. Assessing T cell responses to potential antigens and the anti-tumoral activities of stimulated T cells in vitro in order to select therapeutic targets before clinical trials will be a crucial individualized approach to overcome these limitations. Patientderived organoids (PDOs) established with a three-dimensional (3D) culture system modeling the tumoral microenvironment have been demonstrated to preserve tumoral features even after longterm in vitro culture [14,15]. A co-culture system of tumoral organoids and immune cells was found to be useful for optimizing personalized immunotherapies, including chimeric antigen receptor T cell (CAR-T) and adaptive T cell (ACT) therapy [16–20]. Thus, a PDO-based platform provides the possibility of analyzing the anti-tumoral activities of T cells stimulated by the immunogenic peptides derived from circRNAs.
Liver cancer ranks as the second most lethal malignant disease, with few effective therapies [21]. With the aim of exploring therapeutic strategies for hepatobiliary tumors, we herein describe the landscape of tumor antigens derived from circRNAs in organoids, validate the immunogenicity of peptides generated from tumorspecific circRNAs, and discuss the anti-tumoral attack of peptidereactive cluster of differentiation 8 (CD8) T cells.
《2. Materials and methods》
2. Materials and methods
《2.1. Human subjects》
2.1. Human subjects
From July 2018 to October 2020, surgical tumor resection (1–3 cm3 ) and peripheral blood (3–5 mL) were obtained from 27 patients, peritumor samples (1–3 cm3 ) were obtained from 47 patients with a confirmed diagnosis of hepatobiliary cancer, and peripheral blood samples (1–3 mL) were obtained from six healthy donors. The clinical characteristics of the patients are provided in Table S1 in Appendix A, and the human leukocyte antigen (HLA)-A, -B, and -C (HLA-ABC) information of patients and healthy donors is shown in Table S2 in Appendix A. This study was approved by the Ethics Committee of Eastern Hepatobiliary Surgery Hospital (Shanghai, China), and informed consent was obtained.
《2.2. PBMC isolation》
2.2. PBMC isolation
Peripheral blood mononuclear cells (PBMCs) were isolated from peripheral blood by means of Ficoll-Paque separation solution (GE Healthcare, UK) gradient centrifugation and were preserved with CryoStor CS10 medium (STEMCELL Technologies, Canada) in liquid nitrogen for later use.
《2.3. Tumoral organoid culture》
2.3. Tumoral organoid culture
Hepatobiliary tumor organoids were established and cultured as previously described [22]. In brief, small pieces of tumor tissues (0.5–1.5 cm3 ) were digested using a digestion buffer (Dulbecco’s modified Eagle medium (DMEM; Gibco, USA) with 4 mg·mL–1 collagenase D (Roche, Germany), 0.1 mg·mL–1 deoxyribonuclease I (DNase I; Sigma-Aldrich, USA), 2 μmol·L–1 Rho-associated kinase inhibitor (Y27632; Sigma-Aldrich), and 100 μg·mL–1 Primocin (InvivoGen, USA) at 37 °C for 30–90 min and filtered with a 70 μm nylon cell strainer (Falcon, USA)). After washing twice with cold advanced DMEM/F12 (Gibco), single cells were resuspended in an organoid culture medium composed of advanced DMEM/F12 with 1% penicillin/streptomycin, 1% GlutaMAX, 10 mmol·L–1 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES), 100 μg·mL–1 primocin, 1:50 B27 supplement (without vitamin A), 1.25 mmol·L–1 N-acetyl-L-cysteine, 50 ng·mL–1 mouse recombinant epidermal growth factor (EGF), 100 ng·mL–1 recombinant human fibroblast growth factor 10 (FGF10), 1 ng·mL–1 recombinant human FGFbasic, 25 ng·mL–1 recombinant human hepatocyte growth factor HGF), 10 μmol·L–1 forskolin, 5 μmol·L–1 transforming growth factor β receptor 1 inhibitor (A8301), 10 μmol·L–1 Y27632, 10 mmol·L–1 nicotinamide, 10% (v/v) Rspo-1 conditioned medium, 30% (v/v) Wnt3a-conditioned medium, and 5% (v/v) Noggin conditioned medium. The cell suspension mixed with cold Matrigel Basement Membrane Matrix (Corning, USA) was cultured in a six well suspension culture plate at 37 °C for 30 min; then, medium was added. Established organoids were passaged by incubating in Tryple Express (Gibco) for 5 min at 37 °C; dissociated single cells were cultured in fresh medium-matrix approximately every week. Organoids later than passage 5 and earlier than passage 30 were used in this study.
《2.4. Sequencing analysis》
2.4. Sequencing analysis
Organoids were harvested and dissociated into single cells with Tryple Express. After being washed twice with phosphate-buffered saline (PBS), a cell pellet was prepared for RNA-sequencing (RNAseq) and whole genome sequencing (WGS). RNA extraction, library construction, RNA-seq, and WGS were performed by Genergy Biotechnology Co., Ltd. (China). The libraries were sequenced using the Illumina NovaSeq 6000 platform following the manufacturer’s instructions (Illumina Inc., USA). The coverage depth was 10–12 Gb in RNA-seq, 240 Gb for tissue and organoids, and 120 Gb for blood in WGS.
《2.5. HLA typing》
2.5. HLA typing
Four-digit HLA class I alleles were identified by Athlates [23], HLA-HD [24], and HLA-VBSeq [25] with WGS data (Genergy Biotechnology Co., Ltd.). The overlapped results of the software were used as the HLA class I alleles of each sample. HLA class I alleles of healthy PBMCs and five organoids used in peptide tests were analyzed by polymerase chain reaction-sequence-based typing (PCR-SBT; CSTB, China).
《2.6. Detection and quantitation of circRNAs》
2.6. Detection and quantitation of circRNAs
Trimmomatic (v0.39)↑ was used to remove adapter sequence fragments and low-quality fragments from the sequencing data. The circRNAs of 27 organoids and 47 peritumor samples were detected by CIRCexplorer2 from the RNA-seq data [26], and the number of junction reads was inputted to detect circRNAs. The total RNA and polyA(–) RNA-seq data of normal cell lines from the ENCODE database↑ , including a total of 145 samples from 61 normal cell lines, were downloaded and were detected as described above. The reference genome hg38 from the University of California at Santa Cruz (UCSC) genome browser , was used for all mapping and subsequent analyses. In order to obtain organoid-specific circRNAs, we excluded circRNAs that could be detected in either normal cell lines or peritumor samples from all circRNAs. Organoid-specific circRNAs were normalized by spliced reads per billion mapping (SRPBM) = (number of junction reads)/(number of mapped reads) × 1 000 000 000.
, was used for all mapping and subsequent analyses. In order to obtain organoid-specific circRNAs, we excluded circRNAs that could be detected in either normal cell lines or peritumor samples from all circRNAs. Organoid-specific circRNAs were normalized by spliced reads per billion mapping (SRPBM) = (number of junction reads)/(number of mapped reads) × 1 000 000 000.
↑ http://www.usadellab.org/cms/index.php?page=trimmomatic.
 https://www.encodeproject.org.
https://www.encodeproject.org.
《2.7. Determine the coding potential of circRNAs》
2.7. Determine the coding potential of circRNAs
IRESfinder was used to predict the IRES element for each circRNA [12]. ORFfinder, which was developed by the National Center for Biotechnology Information, was used to predict the ORF element for each circRNAs.
《2.8. Antigen peptide prediction》
2.8. Antigen peptide prediction
Peptides with 9–11 amino acid length (mer) derived from organoid-specific circRNAs, which were across the backspliced junction and bind to the HLA-ABC alleles, were predicted with the score tool [27]. In general, this method was used to evaluate the immunogenicity of antigen epitopes in terms of proteasomal cleavage and transporter associated with antigen processing (TAP) transport capacity (NetCTLpan 1.1), major histocompatibility complex (MHC) binding affinity (NetMHCpan4.1), and TCR recognition of the peptides. A PepMatch tool was designed for the normal intra-tissue peptides in humans that are the most similar to candidate peptides in the reference protein database (UniProt). The mismatch number measures the sequence difference of peptides against those in the database↑↑. Peptides with high scores and a rank of less than 0.5 derived from each HLA allele were selected for the validation experiment.
↑↑ https://pypi.org/project/pepmatch/.
《2.9. Gene ontology (GO) enrichment analysis》
2.9. Gene ontology (GO) enrichment analysis
We sorted the expression levels of the standardized organoidspecific circRNAs and selected the top genes that produce the most circRNAs. The ClusterProfiler package was used to identify the GO, biological process (BP), cellular component (CC), and molecular function (MF) in the gene list, with adjusted P < 0.05 according to the Benjamini–Hochberg method [28].
《2.10. Peptide synthesis》
2.10. Peptide synthesis
Customized peptides were obtained from GL Biochem (Shanghai) Ltd. and dissolved in dimethyl sulfoxide (DMSO) for experiments.
《2.11. HLA class I peptide purification via co-immunoprecipitation (COIP) and mass spectrometry (MS) analysis》
2.11. HLA class I peptide purification via co-immunoprecipitation (COIP) and mass spectrometry (MS) analysis
The HLA class I peptides were purified by means of CO-IP and MS analysis, as follows: Organoids were harvested and dissociated into single cells with Tryple Express. After being washed twice with PBS, a pellet of PDOs (1 × 107 –2 × 107 cells·pellet–1 ) was lysed in cold Western and IP lysis buffer containing protease inhibitor cocktail (Beyotime Biotechnology, China) with shaking on ice for 40 min. Cell lysates were centrifugated at 4 °C at 12 000 revolutions per minute (rpm) for 10 min. Soluble lysates were incubated with anti-HLA class I antibody (W6/32; Abcam, UK) and protein A/G magnetic beads (MedChemExpress, USA) according to the manufacturer’s instructions. After the beads were washed and eluted, supernatant was prepared for 10% sodium dodecylsulfate polyacrylamide gel electrophoresis (SDS-PAGE). The collected gel pieces were incubated with 50 μL dithiothreitol (DTT) solution (10 mmol·L–1) at 56 °C for 30 min, and then with 50 μL iodoacetamide solution (55 mmol·L–1) at 37 °C in the dark for 10 min. Neat acetonitrile was then added to shrink the gel pieces. After the liquid was removed, the gel pieces were incubated with trypsin buffer (13 ng·μL–1 in 10 mmol·L–1 NH4HCO3 containing 10% acetonitrile) at 37 °C overnight. The digested peptides were extracted from the gel by sequentially adding 0.1% formic acid in 50% acetonitrile, 0.1% formic acid in 80% acetonitrile, and 100% acetonitrile.
The samples were analyzed with online nanospray liquid chromatography with tandem mass spectrometry (LC-MS/MS) on a Q exactive plus mass spectrometer equipped with an EASY-nanoLC 1000 system (Thermo Fisher Scientific, USA) by Kigene Co., Ltd. (China). PEAKS DB (Bioinformatics Solutions Inc., Canada) was set up to search uniprot_Homo_sapiens (version201907, 20 414 entries) and our database of predicted antigen peptides.
《2.12. In vitro co-culture with peptide》
2.12. In vitro co-culture with peptide
Healthy PBMCs were expanded with 25 μL·mL–1 ImmunoCult Human CD3/CD28 T Cell Activator in the ImmunoCult-XF T Cell Expansion Medium supplemented with 10 ngmL1 human recombinant IL-2 (STEMCELL Technologies) at 1 × 106 cells·mL–1 , 37 °C, and 5% CO2 for 3–6 days while changing to fresh medium every three days. A 96-well U-bottom plate was coated with 5 μg·mL–1 of anti-CD28 (clone 28.2, eBioscience, USA) at 4 °C overnight before use. On Day 0, 1 × 105 PBMCs were incubated with 25 μmol·L–1 of peptide (DMSO was used as the negative control) in 200 μL of T cell medium (AIM-V medium supplemented with 10% human AB serum (Gemini, USA), 1% UltraGlutamine (Lonza, Switzerland), 1% penicillin/streptomycin, interleukin (IL)-2 (100 U·mL–1 ; PeproTech, USA), IL-7 (10 ng·mL–1 ; PeproTech), and IL-15 (10 ng·mL–1 ; PeproTech)) in each well, with at least two replicates. On Day 3, the cells were evenly split and 100 μL of T cell medium (2× concentrated) was added. On Day 6, half of the medium, including peptides and cytokines (2× concentrated), was refreshed. Cell culture supernatants were collected for enzyme-linked immunosorbent assay (ELISA) analysis. After three cycles of peptide stimulation, PBMCs were collected and analyzed using flow cytometry.
《2.13. T cell response analyzed by flow cytometry》
2.13. T cell response analyzed by flow cytometry
PBMCs stimulated with the same peptides were collected and stained with FV450 (Becton, Dickinson and Company (BD), USA) for 15 min at room temperature (RT) and then stained with anti CD107a phycoerythrin-cyanine 7 (PE-Cy7), anti CD3 fluorescein isothiocyanate (FITC), anti CD45 allophycocyanin-cyanine 7 (APC-Cy7), anti CD4 PE, and anti CD8 APC (BD) for 30 min at 4 °C. Cells were washed, fixed, and permed using a fixation/permeabilization kit (PeproTech), and then stained with intracellular anti IFNγ BV510 (BioLegend, USA) for 15 min at 4 °C. Cells were washed and analyzed using a Canto II (BD) or CytoFLEX (Beckman Coulter, Inc., USA) flow cytometer.
《2.14. IFNγ ELISA assay》
2.14. IFNγ ELISA assay
The IFNγ in the supernatants was quantified with a human IFNγ high-sensitivity ELISA kit (Abcam) according to the manufacturer’s instructions.
《2.15. Organoid killing assay》
2.15. Organoid killing assay
The PBMCs were incubated for nine days with three cycles of antigen peptide stimulation. After stimulation, the PBMCs were washed and stained with anti CD3 AF700, anti CD8 APC, and propidium iodide (PI; BD). CD3+ CD8+ PI– cells were isolated using fluorescence-activated cell sorting (FACS) for the organoid killing assay. Organoids were dissociated to single cells with Tryple Express, labeled with carboxyfluorescein succinimidyl ester (CFSE; BD), and co-cultured in triplicate with sorted T cells at a 10:1 effector:target ratio. After three days of co-culture, organoids and T cells were collected. Cells were washed and stained with anti CD45 APC-Cy7, Annexin V PE, and 7-actinomicin D (7-AAD; BD) in Annexin V buffer (BD) for 15 min at RT. The apoptosis of organoids labeled with CFSE was analyzed with a Canto II (BD) or CytoFLEX (Beckman Coulter, Inc.) flow cytometer. FITC+ CD45 APC-Cy7– was used to gate the organoids and Annexin V–7-AAD– was used to identify live cells.
《2.16. Statistics》
2.16. Statistics
A statistical analysis was performed with SPSS Statistics 23 (SPSS Inc., USA). All values are presented as the mean ± standard deviation (SD) or standard error of the mean (SEM). Two-tailed paired and unpaired t tests were performed to compare multiple groups. A P < 0.05 was considered to be a statistically significant difference.
《3. Results》
3. Results
《3.1. Hepatobiliary tumor organoids as a feasible model to profile circRNAs》
3.1. Hepatobiliary tumor organoids as a feasible model to profile circRNAs
We have previously established the long-term expansion of patient-derived hepatobiliary tumor organoids (PDHOs) [22]. Here, we validated the performance of PDHOs as a platform with RNAseq analysis for circRNA detection and the immunogenicity assessment of circRNA-derived peptides (Fig. 1(a))↑ . Using 61 human normal cell lines and 47 peritumor tissues as controls, we detected 7465 tumor-specific circRNAs across 27 PDHOs (Fig. 1(b)). To further characterize these tumor-specific circRNAs, we compared the features and found that the circRNAs were enriched with chromosome (chr) 1. We further calculated the relative amount of circRNAs per megabase (Mb) of the chromosomes and found that chr 17 was an extremely active region in producing circRNAs (Fig. 1(c)). Previous studies have shown that numerous crucial mutations are on chr 17, including breast cancer 1 (BRCA1) and tumor protein P53 (TP53), implying the essential role of this chromosome in the tumor-specific circRNA generation of hepatobiliary cancer. The number of one-exon-derived circRNAs was much higher than others (Fig. 1(d)). The exon length of the circulars increased with the number of exons per circular, except for circRNAs from one single exon (Fig. 1(e)). Further effort is required to understand the essential role of one-exon-derived circRNAs in hepatobiliary cancer.
↑ Elements provided by https://biorender.com/.
Focusing on the circRNAs shared by different PDHOs, we found that 15 circRNAs were shared by more than 4 in 27 PDHOs. Dynein axonemal heavy chain 14 (DNAH14)_chr1_2250- 38693_225051795_+, which was detected in six PDHOs, showed the widest distribution, followed by AFP_chr4_ 73450614_73455290_+ (5 in 27) (Fig. 1(f)). DNAH14 has also been reported as the top host gene of the circRNAs in pancancer studies, suggesting that PDHOs retain the circRNA features of primary tissues [29].
In our analysis, most parental genes (over 99%) were able to generate fewer than ten circRNAs; notably, 39 circRNAs originated from the Pals1-associated tight junction gene (PATJ) with the highest number of circRNAs from the parental gene (Fig. 1(g)). Among the top 30 oncogenes with high relevance scores ,17 oncogenes were predicted to generate circRNAs, including ataxia telangiectasia mutated gene (ATM), which was predicted to generate 11 circRNAs, and breast cancer 2 gene (BRCA2) and BRCA1, which respectively ranked as the first and second oncogenes in terms of gene’s relevance score on the oncogene list (Fig. 1(g)). Due to the close relationships between circRNAs and their parental genes [30], we performed a GO function enrichment analysis of the top 20 high-expressing circRNA-related host genes. The results of the GO enrichment analysis showed that 16 GO BP, one GO CC (nuclear pore nuclear basket), and no GO MF terms were significantly enriched (adjusted P < 0.05, Fig. 1(h)). GO enrichment showed that three genes were enriched in lysosomal transport, mRNA transport, vacuolar transport, nucleic acid transport, RNA transport, and the establishment of RNA localization (P = 0.028), indicating the important underlying functions of transport in circRNA-related tumorigenesis and development.
,17 oncogenes were predicted to generate circRNAs, including ataxia telangiectasia mutated gene (ATM), which was predicted to generate 11 circRNAs, and breast cancer 2 gene (BRCA2) and BRCA1, which respectively ranked as the first and second oncogenes in terms of gene’s relevance score on the oncogene list (Fig. 1(g)). Due to the close relationships between circRNAs and their parental genes [30], we performed a GO function enrichment analysis of the top 20 high-expressing circRNA-related host genes. The results of the GO enrichment analysis showed that 16 GO BP, one GO CC (nuclear pore nuclear basket), and no GO MF terms were significantly enriched (adjusted P < 0.05, Fig. 1(h)). GO enrichment showed that three genes were enriched in lysosomal transport, mRNA transport, vacuolar transport, nucleic acid transport, RNA transport, and the establishment of RNA localization (P = 0.028), indicating the important underlying functions of transport in circRNA-related tumorigenesis and development.
 The oncogene list and each gene’s relevance score are publicly available at https:// www.genecards.org/.
The oncogene list and each gene’s relevance score are publicly available at https:// www.genecards.org/.
《Fig. 1》

Fig. 1. Tumor-specific circRNAs are detectable in hepatobiliary tumor organoids. (a) Flow chart outlining the antigen peptide prediction, selection, and validation. (b) Venn diagram comparing the circRNAs from organoids, peritumor tissues, and normal cell lines. (c) Number of circRNAs from each chromosome (amount of circRNAs per Mb of the chromosomes shown with black dots). (d) Amount of circRNAs generated by different numbers of exons. (e) Exon length of circRNAs generated by different numbers of exons (two-tailed unpaired t test, ***: P < 0.001). (f) Scatterplot of top-frequency circRNAs in organoids (dot size represents the expression level of circRNAs; dot color represents the number of organoids with the same circRNAs). HCC: hepatocellular carcinoma. (g) Proportion of different circRNA numbers derived from the same host genes. BIRC6: baculoviral inhibitor of apoptosis protein (IAP) repeat containing 6. (h) GO analysis of the parental genes of the top 20 high-expressing circRNAs.
《3.2. CircRNAs predicted to generate antigen peptides》
3.2. CircRNAs predicted to generate antigen peptides
Among a total of 7815 tumor-specific circRNAs across 27 PDHOs, 5937 circRNAs were predicted to contain IRES; moreover, 5130 circRNAs across the junction had the potential to encode proteins, accounting for 65.64% of all tumor-specific circRNAs (Fig. 2(a)). A previously published score tool [27] was used to quantitatively assess the immunogenic potential of the peptides. The immunogenic-related scores of 18 971 predicted translatedcircRNA-derived peptides of 27 PDHOs were ranked (mean: 0.037, range: 0–0.954) (Fig. 2(b)). The results of the GO enrichment analysis of the host genes generating the top ten circRNA-derived peptides showed that 23 GO BP, seven GO CC, and nine GO MF terms were significantly enriched (adjusted P < 0.05). Two genes were related to guanyl-nucleotide exchange factor activity, GTPase activator activity, GTPase regulator activity, and nucleosidetriphosphatase regulator activity (P = 0.0426, Fig. 2(c)), indicating their potential roles in the formation of tumor antigens. Consistent trends in the translated tumor-specific circRNA count and circRNAderived antigen load (counts of predicted circRNA-derived antigen peptides) in individuals are shown in Fig. 2(d); however, neither circRNA count nor circRNA-derived antigen load was consistent with the tumor mutational burden (TMB). In a further correlation analysis, a significant positive correlation was found between circRNA count and circRNA-derived antigen load, but no significant positive correlation was observed between TMB and circRNAderived antigen load (Figs. 2(e) and (f)), implying that the circRNA-derived antigen load was not related to TMB but to the circRNA burden. For cancer patients with a lack of nonsynonymous mutation-derived neoantigens, circRNA-derived antigens may offer potential therapeutic targets. Neoantigen load has been reported to be related to the prognosis of hepatocellular carcinoma (HCC) [31]. When comparing the patients at different tumor-node-metastasis (TNM) stages, we found that the circRNAderived antigen load was relatively lower in patients with a later tumor stage (Fig. 2(g)). Unfortunately, no significant difference was found, possibly due to the limited case size.
《Fig. 2》

Fig. 2. Correlations between TMB/circRNA-derived antigen load and tumor stage. (a) Venn diagram comparing the counts of total tumor-specific circRNAs, circRNAs with IRES, and circRNAs with the ORF across the junction. (b) Immunogenicity-related score rank for all 18 971 predicted peptides (peptides are ordered by scores). (c) GO analysis of the parental genes of the top ten circRNAs generating antigen peptides. (d) Counts of translated tumor-specific circRNAs, circRNA-derived antigen peptides, and TMBs of 27 organoids. (e) Positive correlations between circRNA count and circRNA-derived antigen load (estimated by liner regression). (f) Positive correlations between TMB and circRNA-derived antigen load (estimated by liner regression). (g) CircRNA-derived antigen load correlated with tumor stage (two-tailed unpaired t test). Data are presented as mean ± SEM.
《3.3. Hepatobiliary tumor organoids profiling the circRNA-derived HLA class I antigen landscape》
3.3. Hepatobiliary tumor organoids profiling the circRNA-derived HLA class I antigen landscape
To characterize the translated tumor-specific circRNAs with predicted potential to generate antigen peptides (termed AgcircRNAs), we analyzed the features and found that chr 17, which generated the largest amount of Ag-circRNAs per Mb, was an active region in producing both circRNAs and Ag-circRNAs (Figs. 1(c) and 3(a)). Ag-circRNAs and the remaining non-Ag-circRNAs were then compared; we found that the chromosome distributions of the Ag-circRNAs’ and non-Ag-circRNAs’ host genes were different, as were the numbers of exons generating Ag-circRNAs or non-AgcircRNAs. Ag-circRNAs were enriched from chr 2, while non-AgcircRNAs were enriched from chr 1 (Fig. 3(b)). Ag-circRNAs were mainly generated from three, four, or one exon, while the number of one-exon-derived non-Ag-circRNAs was extremely high (Fig. 3(c)). Similar to the circRNAs shown in Fig. 1(e), the exon length of the Ag-circRNAs increased with the number of exons per circular, except for the Ag-circRNAs from one single exon (Fig. 3(d)). Detailed comparisons showed that the single-exon and over-20-exon length of Ag-circRNAs was significantly longer than those of non-Ag-circRNAs (Fig. 3(e)). These differences reveal several distinct features of Ag-circRNAs, which help to optimize the distinguishment of Ag-circRNAs in hepatobiliary cancer.
《Fig. 3》

Fig. 3. Characteristics of Ag-circRNAs in hepatobiliary tumor organoids. (a) Number of Ag-circRNAs (translated tumor-specific circRNAs with the predicted potential to generate antigen peptides) from each chromosome are shown; amounts of Ag-circRNA per Mb of chromosomes are shown with black dots. (b) Number of Ag-circRNAs and non-Ag-circRNAs from each chromosome, ordered by the number of Ag-circRNAs. (c) Amount of Ag-circRNAs and non-Ag-circRNAs generated by different numbers of exons, ordered by the number of Ag-circRNAs. (d) Exon length of Ag-circRNAs generated by different numbers of exons (two-tailed unpaired t test, ***: P < 0.001). (e) Comparison of exon length of Ag-circRNAs and non-Ag-circRNAs generated by different numbers of exons (two-tailed unpaired t test, ***: P < 0.001).
Building a tumor antigen peptide library shared by individuals is essential in order to offer timely and effective treatments to cancer patients. The high-frequency Ag-circRNAs are shown in Fig. 4(a). Twenty-one Ag-circRNAs were observed in over three of 27 organoids, including circDNAH14 (six organoids carrying circDNAH14). Notably, circDNAH14 also ranked as the top highfrequency circRNA in the tumor-specific circRNA analysis shown in Fig. 1(f).
We evaluated all 18 971 predicted peptides derived from 3950 circRNAs to reveal the features of circRNA-derived antigen peptides. We found that candidate peptides were enriched in the 9 mer subset, but we found no obvious difference in the distribution of HLA-ABC alleles (Fig. 4(b)); moreover, the immunogenicrelated scores of 11 mer peptides and HLA-A binding peptides were the highest by a significant margin (Fig. 4(c)).
《Fig. 4》

Fig. 4. Features of predicted circRNA-derived antigen peptides. (a) Top-frequency Ag-circRNAs. (b) Proportions and counts of predicted circRNA-derived antigen peptides with each HLA allele and length for individual organoids. (c) Immunogenicity-related scores of predicted circRNA-derived antigen peptides with a comparison of each HLA allele and length (two-tailed unpaired t test). Data are presented as mean ± SD.
《3.4. CircRNA-derived HLA-ABC-presented peptides confirmed with MS》
3.4. CircRNA-derived HLA-ABC-presented peptides confirmed with MS
To evaluate the potential of the circRNAs to generate antigen peptides, we analyzed five PDHOs individually. In total, 1609 circRNAs were identified as tumor specific using peritumor tissues and normal cell lines as the controls; 568 circRNAs with no detectable translation ability (i.e., that do not encode proteins) were further excluded. On average, 698 9–11 mer HLA-ABCbinding peptides derived from 156 translated tumor-specific circRNAs of organoids were predicted (Fig. 5(a)). These candidate antigen peptides were then screened with an MS-based proteomics analysis [32], and 13 peptides were detected and identified as HLA-ABC-presented peptides in three out of five of the PDHOs analyzed (Figs. 5(a) and (b) and Fig. S1 in Appendix A). Trypsin digestion induced the detected peptide enriched with lysine–arginine (KR) termini in the MS analysis. An optimized method for protein in-gel digestion for MS identification should be developed in future. Our data indicate that these circRNAderived encoded peptides exhibiting a relatively high affinity with HLA class I molecules show that circRNAs are a potential antigen source.
《Fig. 5》

Fig. 5. Prediction and confirmation of HLA-ABC-presented peptides. (a) Numbers of total, translated, and Ag-circRNAs in five cases; numbers of predicted HLA-ABC binding 9– 11 mer peptides derived from translated circRNAs and from the MS-detected peptide database are shown. (b) Representative HLA-ABC-presented peptides confirmed with MS. FFKLAEHREVR: phenylalanine–phenylalanine–lysine–leucine–alanine–glutamic acid–histidine–arginine–glutamic acid–valine–arginine; NLKLPKVNVWR: asparagine– leucine–lysine–leucine–proline–lysine–valine–asparagine–valine–tryptophan–arginine.
《3.5. Immunogenicity validation of antigen peptides derived from circRNAs》
3.5. Immunogenicity validation of antigen peptides derived from circRNAs
To validate the immunogenicity of the candidate circRNAderived antigen peptides, we assessed the T cell response under peptide stimulation and the anti-tumoral activity of the peptidestimulated T cells (Fig. 6(a))↑ . The peptides detected by MS and/or those having high scores with a rank less than 0.5 in the algorithm-based antigen peptide prediction were selected for validation experiments (Table S3 in Appendix A). The immunogenic potential-related scores of the peptides selected for validation experiments and of paired wild-type peptides are shown in Table S4 in Appendix A. IFN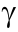 intracellular expression, IFN
intracellular expression, IFN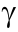 secretion into co-culture supernatants, and the degranulation marker CD107a upon stimulation were evaluated to assess the immunogenicity of peptides [33]. An over 1.5-fold higher frequency of the CD107a+ IFN
secretion into co-culture supernatants, and the degranulation marker CD107a upon stimulation were evaluated to assess the immunogenicity of peptides [33]. An over 1.5-fold higher frequency of the CD107a+ IFN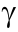 + cells in CD8+ T cells was induced by lysine–leucine–proline– lysine–valine–asparagine–valine–tryptophan–arginine (KLPKVNVWR, HCC-27 peptide 3), compared with the controls treated with DMSO (Fig. 6(b) and Fig. S2 in Appendix A). The raw CD107a+ IFN
+ cells in CD8+ T cells was induced by lysine–leucine–proline– lysine–valine–asparagine–valine–tryptophan–arginine (KLPKVNVWR, HCC-27 peptide 3), compared with the controls treated with DMSO (Fig. 6(b) and Fig. S2 in Appendix A). The raw CD107a+ IFN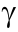 + proportions of the peptide-reactive CD8 T cells are shown in Table S5 in Appendix A. The relatively higher IFN
+ proportions of the peptide-reactive CD8 T cells are shown in Table S5 in Appendix A. The relatively higher IFN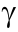 secretion measured by the ELISA consistently supports the tumor-killing potential of the peptides with the highest stimulation of CD107a+ IFN
secretion measured by the ELISA consistently supports the tumor-killing potential of the peptides with the highest stimulation of CD107a+ IFN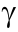 + co-expression T cells (Fig. 6(c)). Interestingly, peptide 3 of HCC-20 and peptide 4 of HCC-27 exhibited the strongest immunogenicity in four of five individuals tested, implying that these two peptides may contribute to the shared antigen peptide library.
+ co-expression T cells (Fig. 6(c)). Interestingly, peptide 3 of HCC-20 and peptide 4 of HCC-27 exhibited the strongest immunogenicity in four of five individuals tested, implying that these two peptides may contribute to the shared antigen peptide library.
↑ Elements used provided by https://biorender.com/.
《3.6. Tumor organoids killed by peptide-reactive CD8 T cells》
3.6. Tumor organoids killed by peptide-reactive CD8 T cells
To evaluate whether peptide-reactive T cells are able to eradicate tumor cells, we performed killing assays using peptides with the strongest stimulation of CD107a+ IFN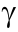 + CD8+ T cells and PDHOs as targets (Fig. 6(a)). The peptide-reactive T cells decreased the proportion of live PDHOs (Fig. 6(d)). Notably, 38% of the surviving HCC-27 PDHOs under DMSO treatment were killed by peptide 4 tyrosine–glycine–phenylalanine–asparagine–glutamic acid–isoleucine–leucine–lysine–lysine (YGFNEILKK)-stimulated CD8 T cells, with only 36% of the PDHOs being left alive, which demonstrates the anti-tumor therapeutic value of targeting circRNA-derived tumor antigens (Fig. 6(e)). The low viability of the organoids in the controls implies that the co-culture condition is not perfect for organoids. Thus, the co-culture system should be optimized in future. Collectively, the peptide-reactive T cells’tumor-killing effect confirms the potential of circRNAs as a source of tumor antigens and the feasibility of using our PDHO-based platform to distinguish immunogenic antigen peptides.
+ CD8+ T cells and PDHOs as targets (Fig. 6(a)). The peptide-reactive T cells decreased the proportion of live PDHOs (Fig. 6(d)). Notably, 38% of the surviving HCC-27 PDHOs under DMSO treatment were killed by peptide 4 tyrosine–glycine–phenylalanine–asparagine–glutamic acid–isoleucine–leucine–lysine–lysine (YGFNEILKK)-stimulated CD8 T cells, with only 36% of the PDHOs being left alive, which demonstrates the anti-tumor therapeutic value of targeting circRNA-derived tumor antigens (Fig. 6(e)). The low viability of the organoids in the controls implies that the co-culture condition is not perfect for organoids. Thus, the co-culture system should be optimized in future. Collectively, the peptide-reactive T cells’tumor-killing effect confirms the potential of circRNAs as a source of tumor antigens and the feasibility of using our PDHO-based platform to distinguish immunogenic antigen peptides.
《Fig. 6》
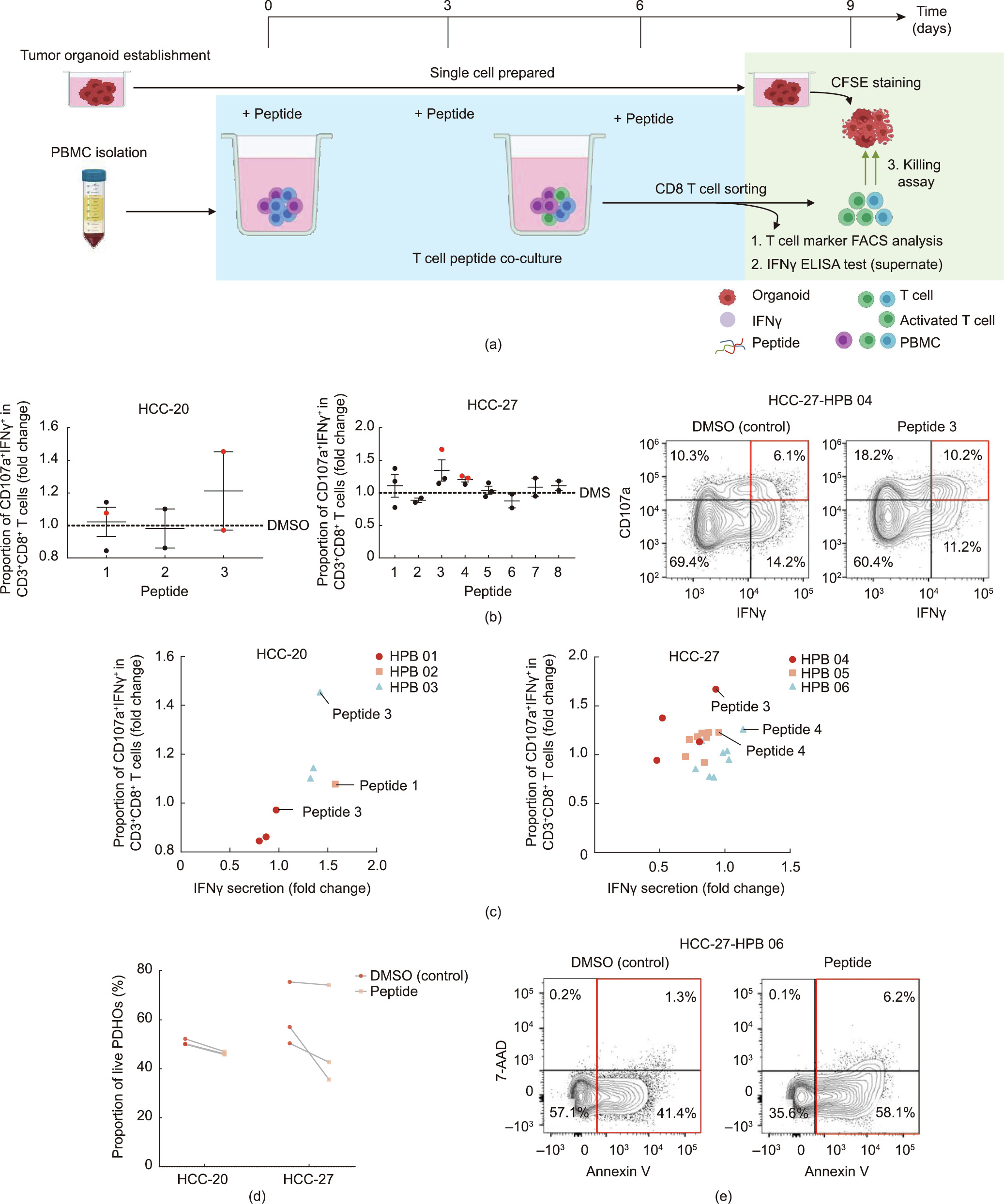
Fig. 6. Identification of immunogenic peptides by T cell peptide co-culture and tumor organoids killed by peptide-reactive T cells. (a) Workflow of validation experiment. After PBMCs were stimulated with peptides for three cycles, T cell activation markers and IFN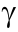 secretion were assessed using flow cytometry and ELISA. Anti-tumoral effects of peptide-reactive CD8+ T cells were evaluated by killing assays after another three days of co-culture with organoids. (b) DMSO stimulation was used as the control, and the fold changes of the three-cycle peptide stimulation-induced CD107a+ IFN
secretion were assessed using flow cytometry and ELISA. Anti-tumoral effects of peptide-reactive CD8+ T cells were evaluated by killing assays after another three days of co-culture with organoids. (b) DMSO stimulation was used as the control, and the fold changes of the three-cycle peptide stimulation-induced CD107a+ IFN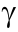 + co-expression of CD3+ CD8+ T cells were quantified (dots indicate independent experiments with different individuals; immunogenic peptides are marked with red). Data are presented as mean ± SEM. Representative flow cytometry plots gated on CD3+ CD8+ T cells. (c) DMSO stimulation was used as the control, and the fold changes of the three-cycle peptide stimulation-induced IFNc secretion of CD3+ CD8+ T cells were quantified (dots indicate independent experiments with different individuals; immunogenic peptides judged by CD107a+ IFN
+ co-expression of CD3+ CD8+ T cells were quantified (dots indicate independent experiments with different individuals; immunogenic peptides are marked with red). Data are presented as mean ± SEM. Representative flow cytometry plots gated on CD3+ CD8+ T cells. (c) DMSO stimulation was used as the control, and the fold changes of the three-cycle peptide stimulation-induced IFNc secretion of CD3+ CD8+ T cells were quantified (dots indicate independent experiments with different individuals; immunogenic peptides judged by CD107a+ IFN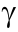 + co-expression are annotated). (d) Organoid killing effects of peptidereactive T cells were assessed according to the proportion of live cells with flow cytometry. The viability of organoids co-cultured with DMSO was used as the control. (Lines present different individuals.) (e) Representative flow cytometry plots gated on organoids. Apoptotic cells (Annexin V+ ) are marked with a red box. HPB: healthy peripheral blood.
+ co-expression are annotated). (d) Organoid killing effects of peptidereactive T cells were assessed according to the proportion of live cells with flow cytometry. The viability of organoids co-cultured with DMSO was used as the control. (Lines present different individuals.) (e) Representative flow cytometry plots gated on organoids. Apoptotic cells (Annexin V+ ) are marked with a red box. HPB: healthy peripheral blood.
《4. Discussion》
4. Discussion
The establishment of primary liver cancer (PLC) [14] and biliary tract carcinoma (BTC) [15] PDOs has been reported. In the current applications of translational medicine, tumor organoids are mainly used as an in vitro model for anti-tumor drug screening and selection [14,15,34]. Organoid-based tumor antigen studies are rarely reported. Here, we successfully established the long-term expansion of PDHOs [22]. To the best of our knowledge, this study is the first to utilize tumor organoids as a platform to perform circRNA-derived tumor antigen peptide prediction and validation.
Thanks to improved high-throughput RNA-seq technology and bioinformatics algorithms, an increasing number of circRNAs are being reliably identified [35]. Accumulating evidence demonstrates that highly stable circRNAs with aberrant cancerous expressions that contribute to tumorigenesis and tumor metastasis are promising therapeutic targets [7,36,37]. According to our RNAseq analysis, out of 41 282 detected circRNAs, 5130 were judged to be translated exonic tumor-specific circRNAs in 27 established organoids; this result indicates the feasibility of the organoid model used in this circRNA study.
Hundreds of antigen peptides have been confirmed across various tumors↑ . Most previous reports have focused on the coding regions of the genome as the source of neoantigens. However, sole mSNV-derived neoantigens may prove to be impractical for downstream clinical use, since they interfere with the low frequency of immunogenic peptides and exhibit limited immunogenicity due to having only one amino acid alteration [3,4]. Emerging interest in developing unconventional tumor antigens has resulted in several studies focusing on the tumor antigen potential of coding regions with aberrant transcription, translation, post-translation, or even non-coding regions [38–43]. According to the definition of tumor antigens, we assume that the translation production of ORFs at the backsplicing junction of tumor-specific circRNA may be a novel source of tumor antigens. To overcome possible central immune tolerance, tumor-specific circRNAs, which are exclusively expressed in tumor tissues and not in peritumor tissues or normal cells, should be preferentially considered as a source of tumor antigens.
↑ Elements provided by https://biorender.com/.
Our analysis of the generated RNA-seq data revealed that 3950 circRNAs possessed the predicted capability to initiate translation and generate tumor antigen peptides. The repertoires of the HLAABC-presented peptides were directly assessed with MS-based immunopeptidomics [44–46]. In our MS analysis, circRNAderived antigen peptides predicted with RNA-seq-based computational algorithms were detected. Despite the defects of the MS method, such as its limited detection sensitivity and its detectability only of peptides with a relatively high affinity to the HLA-ABC [44,45,47], the translation capacity of circRNAs and the presentation ability of circRNA-encoded peptides were still identified in this manner. Aside from short peptides, long peptides derived from circRNAs are worth investigating in future as another antigen source. The peptides presented on the cell surface should be verified through LC-MS/MS.
To overcome the deficiencies of algorithm-based prediction, it is a prerequisite to perform validation experiments on the candidate peptides. The histological architecture, genomic landscape, and expression profiles from the original tumor tissues were demonstrated to be preserved in the organoids [14,15], thereby establishing the desirability of organoids as the attack targets of neoantigen peptide-reactive T cells [18,48,49]. In our study, the quantitative analysis revealed T cell activation (i.e., increased expression of CD107a, IFN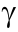 , and IFN
, and IFN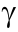 secretion) under immunogenic peptide treatment. The organoid killing assay further confirmed the tumor-elimination ability of the immunogenic peptide-stimulated CD8 T cells. Notably, robust T cell-mediated tumor destruction stimulated by an MS-detected peptide (YGFNEILKK from circTBC1D15) in HCC-27 solidly confirmed the existence and anti-tumoral attack of circRNA-generated antigen peptides. In this way, our organoid-based platform was used to validate circRNAs as a potential source of tumor antigen peptides. More immunogenic circRNA-derived antigen peptides for each patient will be validated in future. The tumor-killing effect of peptide-reactive CD8 cells is likely to be enhanced if stimulated with a combination of immunogenic peptides. This study is limited by its small sample size, so it will be interesting to apply this approach on a large scale to more precisely identify circRNAderived tumor antigens. In future, we will also examine the potential of somatic gene alteration-related circRNAs as a neoantigen source.
secretion) under immunogenic peptide treatment. The organoid killing assay further confirmed the tumor-elimination ability of the immunogenic peptide-stimulated CD8 T cells. Notably, robust T cell-mediated tumor destruction stimulated by an MS-detected peptide (YGFNEILKK from circTBC1D15) in HCC-27 solidly confirmed the existence and anti-tumoral attack of circRNA-generated antigen peptides. In this way, our organoid-based platform was used to validate circRNAs as a potential source of tumor antigen peptides. More immunogenic circRNA-derived antigen peptides for each patient will be validated in future. The tumor-killing effect of peptide-reactive CD8 cells is likely to be enhanced if stimulated with a combination of immunogenic peptides. This study is limited by its small sample size, so it will be interesting to apply this approach on a large scale to more precisely identify circRNAderived tumor antigens. In future, we will also examine the potential of somatic gene alteration-related circRNAs as a neoantigen source.
The distinct features of the predicted antigen-related circRNAs (e.g., being enriched with chr 2 and being generated from three, four, or one exon) and the circRNA-derived antigen peptides with relatively high immunogenicity-related scores (e.g., HLA-A binding and 11 mer length) should be helpful for algorithm alteration to improve prediction performance. It should be noted that the high-frequency antigen-related tumor-specific circRNAs shared by patients, including circDNAH14 (the most common circRNA), require particular focus because they are the most likely to replenish the tumor antigen library.
《5. Conclusions》
5. Conclusions
In conclusion, the PDHOs we present here comprise a practical model for tumor antigen peptide assessment. RNA-seq combined with MS immunopeptidomics was used to predict tumor antigen peptides and to outline the landscape of circRNA-derived antigens in organoids. Cytotoxic activity against tumor organoids triggered by peptide-reactive T cells provided a proof of principle for circRNA-directed T cell immunotherapy. CircRNAs provide a novel source of tumor antigens to be explored as immunotherapeutic targets in clinical applications.
《Acknowledgments》
Acknowledgments
This work was supported by the National Natural Science Foundation of China (U21A20376, 82102871, 81988101, 81903184, 81790633, and 81830054), the Innovation Program of Shanghai Municipal Education Commission (2019-01-07-00-07- E00065), the National Science Foundation of Shanghai (21XD1404600, 21JC1406600, and 22140901000), and the China Postdoctoral Science Foundation (2020M671007).
《Authors’ contribution》
Authors’ contribution
Lei Chen and Peng Wang designed experiments. Hongyang Wang, Dong Gao, Peng Wang, Lei Chen, Wenwen Wang, Lili Ma, Zheng Xing, and Yanjing Zhu developed methods. Jinxia Bao, Yanjing Zhu, Xiaofang Zhao, Yan Zhao, Siyun Shen, Xinyao Qiu, and Shuai Yang collected samples. Wenwen Wang, Tinggan Yuan, Yanjing Zhu, Jinxia Bao, Yali Zong, and Yani Zhang performed experiments. Wenwen Wang, Lili Ma, Zheng Xing, and Tinggan Yuan analyzed data. Wenwen Wang, Lili Ma, and Zheng Xing wrote the manuscript. All authors revised the manuscript and approved the final version. Lei Chen, Peng Wang, Hongyang Wang, and Dong Gao supervised the project.
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Wenwen Wang, Lili Ma, Zheng Xing, Tinggan Yuan, Jinxia Bao, Yanjing Zhu, Xiaofang Zhao, Yan Zhao, Yali Zong, Yani Zhang, Siyun Shen, Xinyao Qiu, Shuai Yang, Hongyang Wang, Dong Gao, Peng Wang, and Lei Chen declare that they have no conflict of interest or financial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2022.06.008.













 京公网安备 11010502051620号
京公网安备 11010502051620号




