《1. Introduction》
1. Introduction
Acute liver failure (ALF) is an unexpected severe consequence of hepatocyte injury. The most prevalent causes of ALF are druginduced liver injury, ischemia, hepatitis viruses, and autoimmunity [1]. Despite its diverse causes, ALF is characterized by similar clinical and pathological features. However, because of its burstiness, it is difficult to study ALF in large and randomized clinical trials [1]. An intriguing finding from animal studies is that pretreatment with oral antibiotics protects the liver from injury, partly due to the limited enteric endotoxin [2,3]; this finding suggests that the gut microbiota plays an important role in liver injury, although the underlying mechanism is still not fully understood.
Emerging studies have demonstrated that the gut microbiota affects almost every aspect of host physiology, and dysbiosis of the gut microbiota has been acknowledged as playing a crucial role in causing and propagating diseases [4,5]. Most of these interactions are driven by a large array of gut microbial components and metabolites. The three kinds of microbial metabolites that are most studied at present are bile acids, short-chain fatty acids (SCFAs), and tryptophan (Trp) metabolites [6].
Germ-free and antibiotic-treated models are commonly used to test the function of the gut microbiota. As an alternative, broadspectrum antibiotic (Abx)-treated mice can avoid some of the complications found in germ-free mice [7]. Abx-induced mice have a broad decrease in bacterial load, which leads to limited exposure to gut microbial components and metabolites, such as lipopolysaccharide (LPS), SCFAs, and secondary bile acids, which affect host susceptibility to diseases [8,9]. Interestingly, gut microbiota depletion leads to high serum Trp, which is observed in both Abx-treated mice and germ-free mice, indicating that the gut microbiota controls Trp availability to the host [10,11]. As an essential amino, Trp follows three metabolic fates, with the production of indoles, kynurenine (Kyn), and serotonin, which are directly or indirectly controlled by the gut microbiota [12]. In addition, the liver plays a central role in the Kyn pathway of the Trp metabolism [13]. Most metabolites of the Kyn pathway, such as Kyn, kynurenic acid, 3- hydroxykynurenine (3-HKYN), and 3-hydroxyanthranilic acid (3- HAA), exert immunosuppressive effects by binding to the aryl hydrocarbon receptor (AhR) [14].
Given that Abx inhibits regulatory T cell (Treg) deficiencymediated lethal inflammation and reduces ischemia reperfusion injury after liver transplantation in mice or humans [15,16], the immunosuppressive and anti-inflammatory effects of Abx, which are probably mediated by the gut microbiota-controlled Trp metabolism, especially the Kyn pathway, need to be further studied. Furthermore, when using Abx-treated mice, the effects of changes in Trp metabolism should be taken into account in order to obtain a comprehensive interpretation.
In the present study, Abx pretreatment for four weeks was found to inhibit the hepatic inflammatory responses and improve the survival of mice with D-(+)-galactosamine hydrochloride (DGal)/lipopolysaccharide (LPS)-induced ALF. Joint analyses of multi-omics data together with mechanistic experiments suggest that this favorable outcome can be partly attributed to the gut microbiota-controlled Trp metabolism and Kyn-dependent AhR activation. This study broadens the current understanding of the role of the gut microbiota–host Trp co-metabolism in ALF, and thus provides a promising target for the better management of ALF.
《2. Materials and methods》
2. Materials and methods
《2.1. Mice》
2.1. Mice
Male C57BL/6 mice (4–6 weeks old) were purchased from Shanghai SLRC Laboratory Animals Co., Ltd. (China). Mice were maintained in an environment with controlled temperature ((25 ± 1) °C) and humidity (60% ± 5%), and were kept on a 12 h:12 h light–dark cycle (light on at 6:00) under specific pathogen-free (SPF) conditions. All mice were fed a standard laboratory diet with free access to food and water. Details of the experimental design are provided in Fig. S1 in Appendix A. The animal protocols were in accordance with the Guide for the care and use of laboratory animals of the National Institutes of Health and approved by the Animal Care Ethics Committee of the First Affiliated Hospital, College of Medicine, Zhejiang University, China.
《2.2. ALF model》
2.2. ALF model
Mice were administered a combination of D-Gal (#G0500, Sigma-Aldrich, Germany) at a dose of 0.75 mg·g-1 of body weight and LPS (Escherichia coli O55:B5, #L2880, Sigma-Aldrich) at a dose of 0.01 μg·g-1 of body weight, which were dissolved in 300 μL sterile phosphate buffered saline (PBS), by intraperitoneal injection once to induce ALF according to published protocols [17].
《2.3. Abx pretreatment》
2.3. Abx pretreatment
Mice were administered a combination of ampicillin sodium salt (1.86 mg per mouse), vancomycin hydrochloride (0.96 mg per mouse), neomycin sulfate (1.86 mg per mouse), and metronidazole (1.20 mg per mouse), which were dissolved in 300 μL sterile distilled water, by oral gavage once per day for four weeks prior to the D-Gal/LPS intraperitoneal injection. All antibiotics were purchased from Sigma-Aldrich.
《2.4. Mucosal barrier degradation》
2.4. Mucosal barrier degradation
Mice were exposed to 2% (w/v) dextran sodium sulfate (DSS; #42867, Sigma-Aldrich) in drinking water for seven days to induce mucosal barrier degradation prior to D-Gal/LPS intraperitoneal injection [18].
《2.5. Probiotic supplement》
2.5. Probiotic supplement
Akkermansia muciniphila was cultured in a 3.7% (w/v) brain heart infusion (BHI; #CM1135, OXOID, USA) medium under an atmosphere of 10% H2, 10% CO2, and 80% N2 at 37 °C in an anaerobic workstation (AW300SG, Electrotek, UK) for 48 h. The bacteria cultures were centrifuged (4 °C, 4000 revolutions per minute (rpm), 5 min), washed three times with sterile PBS containing 2.5% (v/v) glycerol, and resuspended at a final concentration of 1 × 1010 colony forming units (CFU)·mL-1 under anaerobic conditions. Mice were administered 300 μL of suspension containing 3 × 109 CFU Akkermansia muciniphila by oral gavage once per day for four weeks prior to D-Gal/LPS intraperitoneal injection.
《2.6. Fecal microbiota transplant》
2.6. Fecal microbiota transplant
Fresh fecal pellets were collected from the mice that were going to receive a fecal microbiota transplant (FMT) on the day of the FMT. The fecal pellets were weighed and homogenized in sterile PBS (125 mg·mL-1 ), and 150 μL of the suspension was administered to a mouse by oral gavage once straight after the D-Gal/LPS intraperitoneal injection [19]. To protect the anaerobic bacteria, both the preparation and the transplantation processes were operated as soon as possible after the collection of fecal pellets.
《2.7. AhR agonist and antagonist treatments》
2.7. AhR agonist and antagonist treatments
For the AhR agonist treatment, mice were administrated 6- formylindolo[3,2-b]carbazole (FICZ; #SML1489, Sigma-Aldrich) at a dose of 1 μg per mouse by intraperitoneal injection once a week prior to D-Gal/LPS intraperitoneal injection. FICZ was first dissolved in dimethyl sulfoxide (DMSO; #D2650, Sigma-Aldrich) at a final concentration of 0.1 μg·μL-1 ; 10 μL of this solution was then mixed with 190 μL sterile PBS. For the AhR antagonist treatment, mice were administrated 2-methyl-2H-pyrazole-3-carboxylic acid (CH223191, #C8124, Sigma-Aldrich) at a dose of 10 μg per mouse by intraperitoneal injection once a week prior to D-Gal/LPS intraperitoneal injection. CH223191 was first dissolved in DMSO at a final concentration of 1 μg·μL-1 ; 10 μL of this solution was then mixed with 190 μL of sterile PBS [20].
《2.8. Serum biochemical test》
2.8. Serum biochemical test
Serum was extracted by centrifuging the blood samples at 4000 rpm for 10 min. Alanine aminotransferase (ALT), aspartate aminotransferase (AST), and total bilirubin (TBil) levels were measured to evaluate liver function using a standard analyzer (Hitachi 7600-210, Japan).
《2.9. Histology and histological scores》
2.9. Histology and histological scores
Freshly harvested liver tissues were fixed in 4% paraformaldehyde overnight before being dehydrated and embedded in paraffin. Paraffin-embedded liver tissues were sectioned at a thickness of 4 μm, stained with hematoxylin and eosin (H&E), and examined under a biological microscope. The liver histology score is assessed by visualizing the entire H&E-stained section microscopically according to the modified histologic activity index (HAI) [21].
《2.10. Immunofluorescence and immunohistochemistry examination》
2.10. Immunofluorescence and immunohistochemistry examination
For toll-like receptor 4 (TLR4) and F4/80 immunofluorescence staining, after antigen retrieval for the paraffin-embedded sections, the liver sections were simultaneously incubated with anti-TLR4 rabbit antibody (1:500; #GB12186, Servicebio, China) and antiF4/80 mouse antibody (1:500; #GB11027, Servicebio) overnight at 4 °C, and then incubated with Cy3-conjugated goat anti-rabbit antibody (1:300; #GB21303, Servicebio) and Alexa Fluor® 488- conjugated goat anti-mouse antibody (1:400; #GB25301, Service-bio) for 50 min at room temperature. For AhR immunofluorescence staining, after antigen retrieval for the paraffin-embedded sections, the liver sections were incubated with anti-AhR rabbit antibody (1:100; #ab84833, Abcam, UK), and then incubated with Cy3- conjugated goat anti-rabbit antibody (1:300; #GB21303, Servicebio) for 50 min at room temperature. 4' ,6-Diamidino-2-phenylin dole (DAPI; #G1012, Servicebio) was used to stain the nuclei. PBS and mounting medium were separately used to wash the sections. Images of the stained liver sections were captured using a fluorescence microscope (Eclipse C1, Nikon, Japan). For semi-quantitative analysis for immunofluorescence staining, the integrated fluorescence density was counted in five representative high-power fields per mouse using ImageJ software (version 1.51j8, National Institutes of Health, USA).
For proliferating cell nuclear antigen (PCNA) immunohistochemical staining, after antigen retrieval and blocking for the paraffin-embedded sections, the liver sections were incubated with anti-PCNA mouse antibody (1:2000; #GB13010-1, Servicebio) overnight at 4 °C, and then incubated with horseradish peroxidase (HRP)-conjugated goat anti-mouse antibody (1:200; #GB23301, Servicebio) for 50 min at room temperature. 3,3' - Diaminobenzidine (DAB) regent (#K5007, Dako, Denmark) was applied for color developing, and then hematoxylin was used for nuclear re-staining. PBS and mounting medium were separately used to wash the sections. Images of the stained liver sections were captured using a biological microscope (XSP-C204, COIC, China). For quantitative analysis of PCNA staining, the proportion of PCNA-positive hepatocyte nuclei as the proliferation index was counted in five representative high-power fields per mouse.
《2.11. Bacterial DNA extraction》
2.11. Bacterial DNA extraction
The total bacterial DNA was extracted from snap frozen (at –80 °C) colon contents using QIAamp Fast DNA Stool Mini Kit (#51604, Qiagen, Germany) according to the manufacturer’s instructions. DNA concentration and purity were tested using a NanoDrop 2000 spectrophotometer (Thermo Fisher Scientific, USA), and quality was assessed by means of 1.0% (w/v) agarose gel electrophoresis.
《2.12. 16S rRNA gene sequencing》
2.12. 16S rRNA gene sequencing
The total bacterial DNA extracted from the colon content samples was used for 16S rRNA gene sequencing. In brief, the V3–V4 region of the bacterial 16S rRNA gene was amplified by polymerase chain reaction (PCR), and then the sequencing was performed on an Illumina MiSeq platform (Illumina, USA) according to the manufacturer’s instructions. Sequencing data was analyzed using the Quantitative Insights into Microbial Ecology (QIIME) software package. All reads were assigned to operational taxonomic units (OTUs) at a sequence identity of 97% and classified taxonomically using the Greengenes reference database. The α-diversity was measured using the richness and evenness indexes (Chao1 and Shannon). The β-diversity was estimated using the Bray–Curtis distance matrix, which was used to build the principal coordinates analysis (PCoA). Phylogenetic Investigation of Communities by Reconstruction of Unobserved States (PICRUSt) was used to infer the predicted functional composition of the gut microbiome of each sample, which was represented by Statistical Analysis of Metagenomic Profiles (STAMP).
《2.13. RNA extraction and reverse transcription》
2.13. RNA extraction and reverse transcription
The total RNA was extracted from snap frozen (at –80 °C) liver tissue using an RNeasy Plus Mini Kit (#74134, Qiagen) according to the manufacturer’s instructions. After RNA extraction, reverse transcription was performed using a QuantiTect Reverse Transcription Kit (#205311, Qiagen) according to the manufacturer’s instructions. Total RNA and complementary DNA (cDNA) were stored at –80 °C for further processing.
《2.14. RNA sequencing and real-time quantitative PCR》
2.14. RNA sequencing and real-time quantitative PCR
The total RNA of the liver tissue was sheared to synthesis cDNA for RNA library construction. RNA sequencing was performed on the BGISEQ-500 platform (MGI, China). Real-time quantitative PCR was conducted on the ABI ViiA7 real-time PCR system (Applied Biosystems Life Technologies, USA) with a QuantiFast SYBR Green PCR Kit (#204054, Qiagen) to verify the results of RNA sequencing. Mouse actin beta (ACTB; #B662302-0001; Sangon Biotech Co., Ltd., China) was used as endogenous reference gene primers to normalize the measurements of target genes.
《2.15. Global metabolic profiling》
2.15. Global metabolic profiling
Samples were stored at –80 °C after collection until analysis. The samples were preprocessed according to the previous description. In brief, 100 mg of feces was mixed with 300 μL of precooling methanol (MeOH; #173433, Thermo Fisher Scientific) and 200 mg ceramic beads (1 mm; OMNI, USA). Next, the mix was homogenized using a high-speed blender and centrifuged (4 °C, 12 000 rpm) for 10 min. The supernatant was centrifuged (4 °C, 12 000 rpm, 10 min) again and then filtered using 0.22 μm syringe filters (Millipore, USA) before analysis [22]. Fifty microliters of serum was fully mixed with 150 μL of precooling acetonitrile (#186448, Thermo Fisher Scientific), and then the mix was centrifuged (4 °C, 12 000 rpm) for 10 min. One hundred and fifty microliters of the supernatant was centrifuged (4 °C, 12 000 rpm, 10 min) again before analysis. One hundred milligrams of liver tissue was ground in liquid nitrogen and then mixed with 1 mL of methanol/water (4:1 (v/v); #183205, Thermo Fisher Scientific). The mix was kept on ice for 20 min and then centrifuged (4 °C, 15 000 rpm) for 10 min. The supernatant was centrifuged (4 °C, 15 000 rpm) for 10 min again before analysis [23]. For different sample types, a procedural blank was used to monitor contamination, and a pooled quality control (QC) sample was prepared by mixing 10 μL of each sample.
Global metabolic profiling analysis was conducted using ultraperformance liquid chromatography (UPLC) coupled to a quadrupole Orbitrap hybrid mass spectrometry (MS) system. UPLC was performed on a Dionex UltiMate 3000 RS system with a Hypesil Gold C18 analytical column (2.1 mm × 100 mm, 1.9 μm; Thermo Fisher Scientific) at 35 °C [24]. Water with 0.1% formic acid (v/v) was used as mobile phase A and MeOH with 0.1% formic acid (v/v) was used as mobile phase B under electrospray ionizationpositive (ESI+) mode, and water was used as mobile phase A and MeOH was used as mobile phase B under electrospray ionization-negative (ESI–) mode. Before sampling, the column was eluted by 98% mobile phase A and 2% mobile phase B (v/v) at a flow rate of 400 μL·min-1 for 30 min. During sample processing, the linear elution gradient was optimized as follows: For feces samples, the composition of mobile phase B was 2% in 0.5 min, linearly increased to 40% in the next 7.5 min, reached 98% in an additional 7 min and remained there for 3 min, and then decreased to 2% in 2 min before the run stop. For liver tissue and serum samples, the composition of mobile phase B was 2% in 0.5 min, linearly increased to 50% in the next 4.5 min, reached 98% in an additional 5 min and remained there for 5 min, and then decreased to 2% in 3 min before the run stop.
A Q Exactive HF-X MS with heated-ESI-II (HESI-II) ion source (Thermo Fisher Scientific) was used to perform the MS in ESI+ and ESI– mode as previously described [24]. The MS spray voltages were 3.5 kV. The capillary temperature was set at 320 °C with the auxiliary gas at 10 arbitrary units (AU) and the sheath gas at 40 AU. The acquisition mode was full MS with a scan range from 70 to 1050 m/z followed by a data-dependent tamden MS (dd-MS2 ). The resolution was set at 60 000 for the full MS and 15 000 for the dd-MS2 . The MS2 analysis was conducted with the collision energy at 20, 40, and 60 eV.
Before analyzing the samples, five QC samples were run to equilibrate the detection system. During sample analysis, one QC sample was run every ten sample injections to ensure data quality. The top 100 compounds in the blank sample were excluded.
The raw data were collected by means of Xcalibur 4.1 software (Thermo Fisher Scientific) and initially processed by Compound Discoverer 3.1 software (Thermo Fisher Scientific) according to the manufacturer’s guide. In brief, the initial data underwent the Select Spectra, Align Retention Times, and Detect Unknown Compounds nodes. Based on the molecular weight and retention time, compounds were grouped in the Group Unknown Compounds node. The missing chromatographic peaks were filled by the Fill Gaps node. The Background Compounds node and the Normalize Areas node were used for background subtraction and area normalization, respectively. The Predict Compositions node was used for compound identification, and unknown compounds were searched in the in-house database (mass lists and mzVault) and online database (ChemSpider chemical structure sores and mzCloud spectral library).
《2.16. Statistical analysis》
2.16. Statistical analysis
In this study, multiple mice were analyzed as biological replicates. Bar graphs were expressed as mean ± standard deviation (SD) or standard error of the mean (SEM). GraphPad Prism 8.01 (GraphPad Software, Inc., USA) was applied for statistical analyses and graph preparation. The Kolmogorov–Smirnov test was used to verify whether the entire dataset was normally distributed. For datasets that failed normality tests, nonparametric tests were used to analyze differences. For comparisons between two groups, significance was determined by an unpaired Student’s t test or Wilcoxon rank-sum test. For comparisons among more than two groups, an analysis of variance (ANOVA) test or Kruskal–Wallis test was used. An F or Bartlett’s test was performed to determine the differences in variances for the t test and ANOVA, respectively. An unpaired Student’s t test with Welch’s correction or Brown– Forsythe and Welch ANOVA tests were applied when the variances were not equal. Survival time was analyzed by a Kaplan–Meier curve, and p values were assessed with a log-rank (Mantel–Cox) test. Differences were noted as significant at p < 0.05.
《3. Results》
3. Results
《3.1. Antibiotics pretreatment improved the outcome of ALF mice》
3.1. Antibiotics pretreatment improved the outcome of ALF mice
ALF was successfully modeled by D-Gal/LPS intraperitoneal injection, which was verified by increased serum ALT, AST, and TBil levels; extremely high mortality; and massive diffuse liver necrosis in ALF mice compared with control (Con) mice (Fig. 1). In addition, the survival curve of the ALF mice was similar to that in a previous study [17].
Abx pretreatment significantly improved the survival time and survival rate of ALF mice (Fig. 1(b)), whereas other interventions neither benefited nor worsened the prognosis of ALF mice (Fig. S2 in Appendix A). Despite the improved survival, there was no statistical difference in liver function between ALF mice and ALF + Abx mice four hours after D-Gal/LPS injection and, unlike ALF mice, serum AST and TBil levels only showed downward trends in ALF + Abx mice (Fig. 1(a)), which may be due to the timing of blood collection or the individual difference. In regard to histology, H&E staining revealed severe diffuse necrosis and hemorrhage in ALF mice, but few hepatic lesions were present in the surviving ALF + Abx mice (Fig. 1(c)). An analysis of the modified HAI showed a markedly increased score in ALF mice, which was partly decreased in ALF + Abx mice, in comparison with Con mice (Fig. 1(d)).
《Fig. 1》
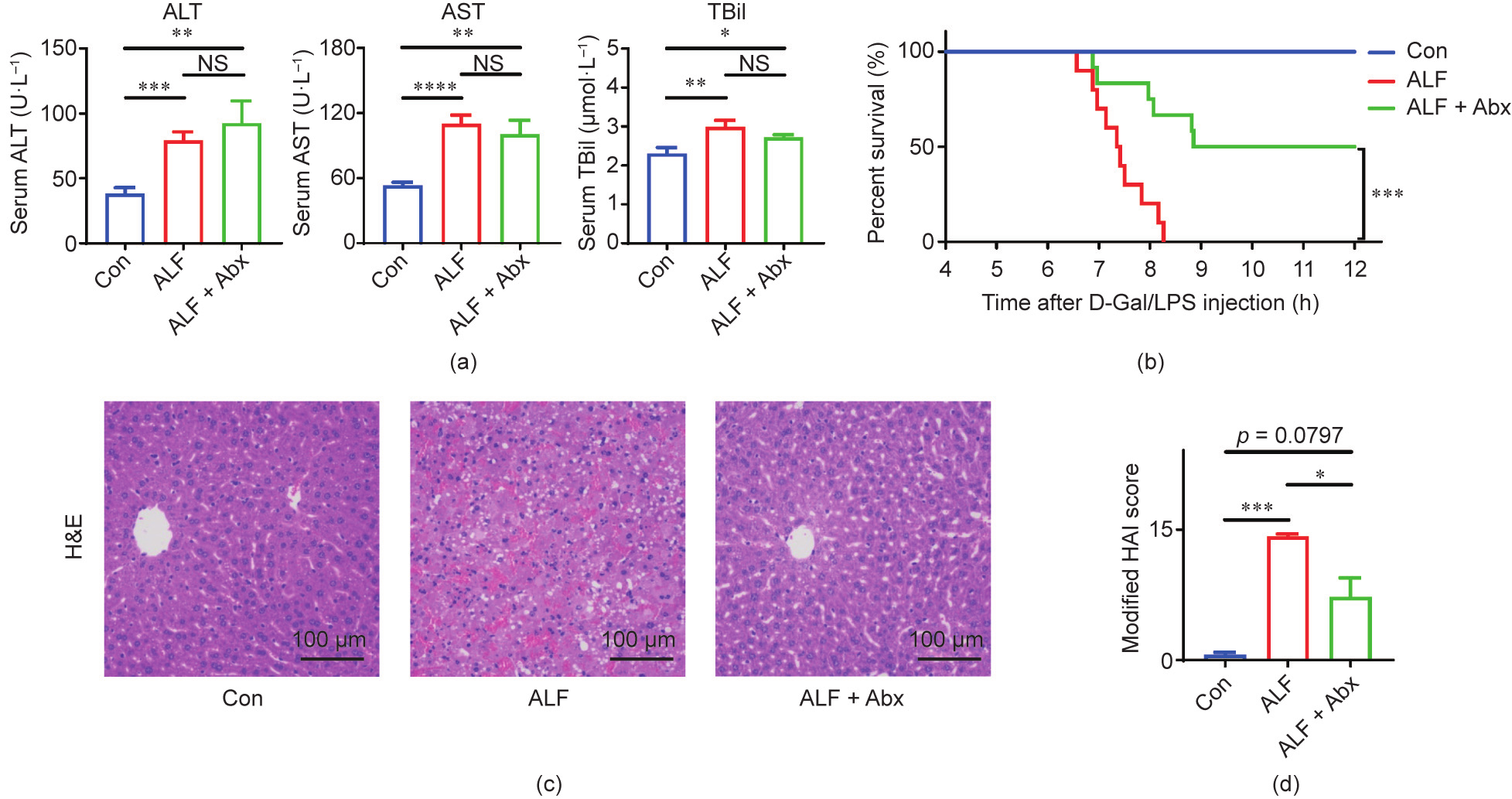
Fig. 1. Antibiotics pretreatment improved the outcome of ALF in mice. (a) Serum ALT, AST, and TBil levels (n = 10–12 per group). (b) Cumulative survival analysis was determined with a Kaplan–Meier curve (n = 10–12 per group). (c) Representative images of hepatic H&E staining. (d) Modified HAI scores of liver histopathology (n = 10–12 per group). Data is shown by mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using ANOVA, Kruskal–Wallis, or log-rank test. NS: no significance.
《3.2. Antibiotics pretreatment changed the hepatic transcriptome of ALF mice》
3.2. Antibiotics pretreatment changed the hepatic transcriptome of ALF mice
The effects of Abx pretreatment on the liver were investigated by comparing the gene expression using RNA sequencing. Principal component analysis (PCA) showed that the samples of Con mice and ALF + Abx mice were quite distinct from those of ALF mice, while the samples of ALF + Abx mice were close to those of Con mice (Fig. 2(a)). Venn diagram analysis revealed that there were 6040 differentially expressed genes (DEGs) between Con mice and ALF mice, while there were 5673 DEGs between ALF mice and ALF + Abx mice and only 1346 DEGs between Con mice and ALF + Abx mice (Fig. 2(b)), suggesting that a large number of hepatic genes were differentially expressed in ALF mice, most of which were curbed by Abx in ALF + Abx mice, compared with Con mice.
In regard to the difference in gene expression between ALF mice and ALF + Abx mice, an analysis of a log–log scatter plot of gene expression levels showed an obvious difference between ALF + Abx mice and ALF mice (Fig. 2(c)). A Kyoto Encyclopedia of Genes and Genomes (KEGG) enrichment analysis of the DEGs showed that the upregulated genes in ALF + Abx mice compared with ALF mice were mainly involved in metabolic pathways, and the downregulated genes were associated with inflammatory responses, such as cytokine–cytokine receptor interaction, chemokine signaling pathway, tumor necrosis factor (TNF) signaling pathway, nucleotide-binding oligomerization domain (NOD)-like receptor signaling pathway, interleukin (IL)-17 signaling pathway, and leukocyte transendothelial migration (Fig. 2(d)). These observations suggested that inflammatory responses were inhibited in ALF + Abx mice, which was probably related to the changes in metabolism pathways.
《Fig. 2》
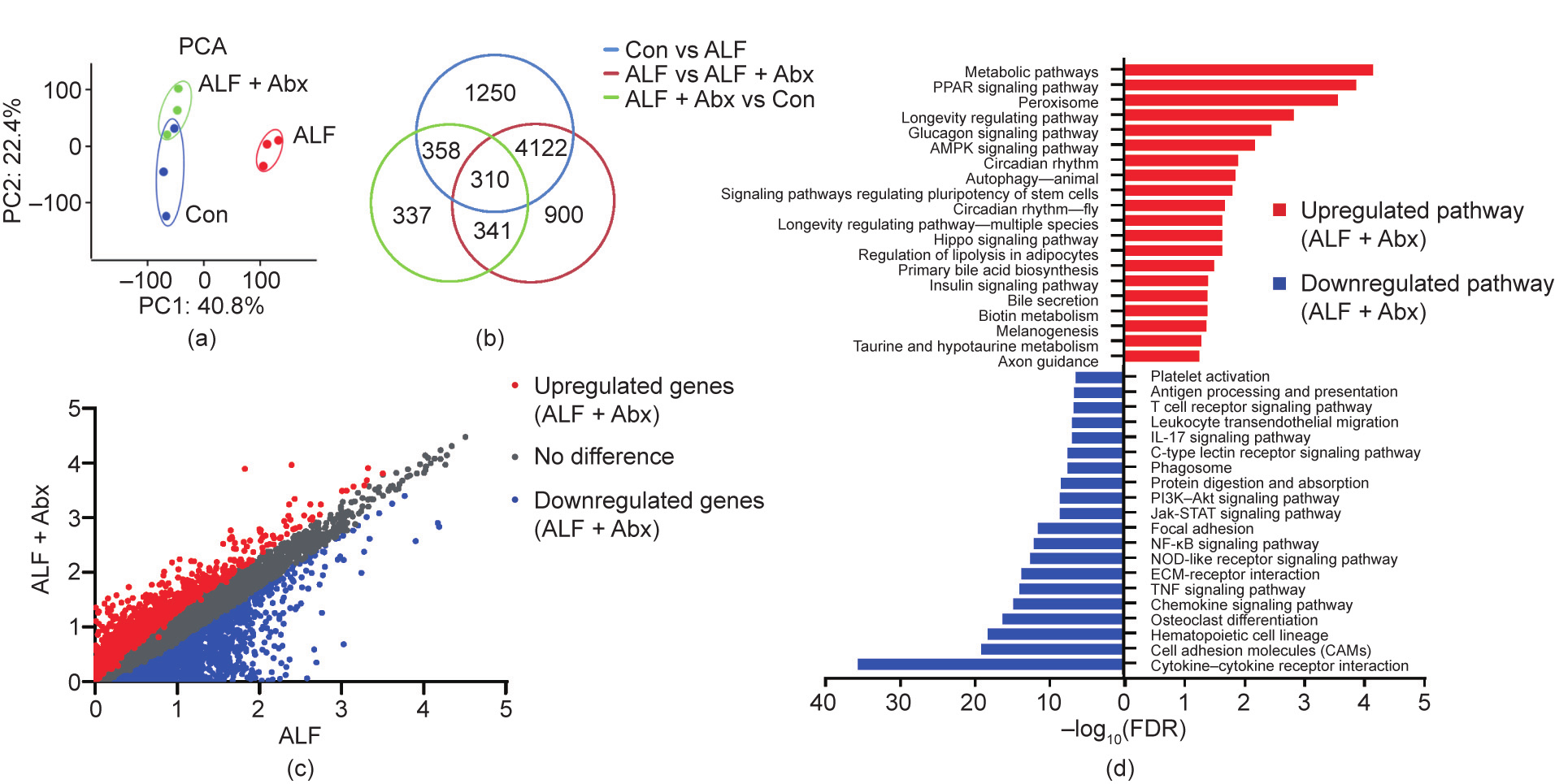
Fig. 2. Antibiotics pretreatment changed the hepatic transcriptome of ALF mice. (a) PCA of the liver RNA sequencing data (n = 3 per group). (b) Venn diagram of DEGs among groups. (c) Log–log scatter plot analysis of gene expression levels between ALF + Abx mice and ALF mice. (d) KEGG pathway enrichment analysis of the DEGs between ALF and ALF + Abx. The top 20 upregulated pathways (red) and the top 20 downregulated pathways (blue) in ALF + Abx compared with ALF are shown. PC: principal component; PPAR: peroxisome proliferator-actiated receptor; AMPK: adenosine monophosphate-activated protein kinase; PI3K: phosphoinositide-3 kinase; ECM: extracellular matrix; FDR: false discovery rate.
《3.3. Antibiotics reduced the bacterial load and diversity while expanding specific taxa》
3.3. Antibiotics reduced the bacterial load and diversity while expanding specific taxa
The effects of Abx pretreatment on ALF indicated that the gut microbiota played a role in this model. Therefore, the composition of the gut microbiota was characterized in order to make sense of the underlying mechanism. Abx treatment for four weeks significantly decreased the broad bacterial load, but did not result in a complete clearance of bacteria (Fig. 3(a)), which can probably be attributed to the emergence of antibiotic-resistant bacteria. The composition of the gut microbiota was identified by 16S rRNA gene sequencing using colon contents. The β-diversity was expressed as the Chao1 and Shannon indexes, which were significantly decreased in ALF + Abx mice compared with Con mice and ALF mice (Fig. 3(b)). Furthermore, the Chao1 index was increased and the Shannon index did not change in ALF mice compared with Con mice. The β-diversity was measured using PCoA with the Bray–Curtis distance (Fig. 3(c)). The gut microbiota composition of ALF + Abx mice was distinct from those of Con mice and ALF mice, while the gut microbiota composition of ALF mice was also distinct from that of Con mice.
At the phylum level, Proteobacteria accounted for the majority of the gut bacteria in ALF + Abx mice, while the relative abundance of Proteobacteria and Firmicutes was increased and the relative abundance of Bacteroidetes was decreased in ALF mice compared with Con mice (Fig. 3(d)). At the genus level, Enterobacteriaceae unclassified, Escherichia–Shigella, and Morganella were the top three bacteria in ALF + Abx mice (Fig. 3(e)).
《Fig. 3》
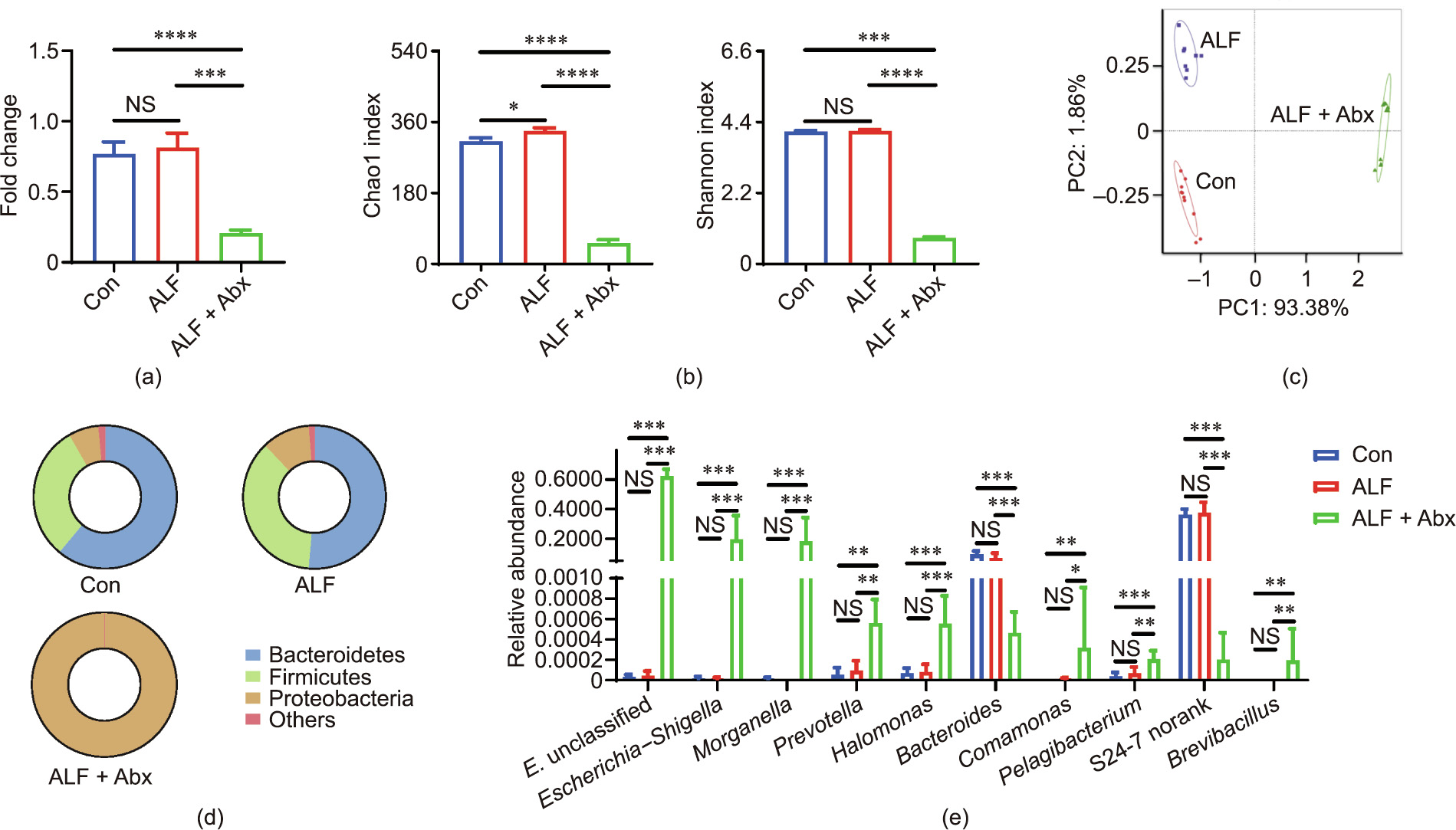
Fig. 3. Antibiotics reduced the bacterial load and diversity while expanding specific taxa. (a) Total bacterial load of the colon content (n = 9–11 per group). (b) Chao1 and Shannon index of the gut microbiota (n = 9–11 per group). (c) PCoA of the gut microbiota composition with Bray–Curtis distance (n = 9–11 per group). (d) Composition of the gut microbiota at the phylum level (n = 9–11 per group). (e) Relative abundance of gut microbiota at the genus level (top 10 in ALF + Abx mice, n = 9–11 per group). Data is shown by mean ± SEM or mean ± SD. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using ANOVA, Kruskal–Wallis, or Wilcoxon rank-sum test. E. unclassified: Enterobacteriaceae unclassified.
Abx reduced the bacterial load and diversity while expanding and collapsing specific indigenous taxa; ALF itself also elicited a compositional shift of the gut microbiota in a short time. Compositional changes will lead to an alteration of the gut microbial function, whose role requires further study.
《3.4. Antibiotics reshaped the gut microbial metabolism with increased Kyn production and reduced Trp excretion》
3.4. Antibiotics reshaped the gut microbial metabolism with increased Kyn production and reduced Trp excretion
In order to assess the functional changes of the gut microbiota, PICRUSt analysis was performed. In regard to the gut microbial metabolism, the mean proportion of the metabolism pathway and the amino acid metabolism pathway were decreased in ALF + Abx mice compared with ALF mice at KEGG level 1 and KEGG level 2, respectively (Figs. S3(a) and (b) in Appendix A). Interestingly, at KEGG level 3 (Fig. 4(a)), several metabolism pathways, including ascorbate and aldarate metabolism, nucleotide metabolism, glutathione metabolism, Trp metabolism, fatty acid metabolism, lipid metabolism, metabolism of cofactors and vitamins, glycan biosynthesis and metabolism, and carbohydrate metabolism, were proportionally increased in ALF + Abx mice compared with ALF mice, and the sphingolipid metabolism was proportionally decreased in ALF + Abx mice compared with ALF mice, indicating that Abx reshaped the gut microbial metabolic phenotype and that the changes in the gut microbial metabolism may be involved in the protective effect of Abx on ALF.
Global metabolic profiling was also performed by UPLC–MS in order to detect gut microbial metabolites. Trp and several Trp metabolites were detected as differential metabolites from the feces of Abx-treated (+) and untreated (–) mice (Fig. 4(b)). In the feces of Abx-treated mice, Trp was decreased, as well as indole3-lactic acid (ILA), indole-3-pyruvic acid (IPYA), and indole-3- acetic acid (IAA), which were the products involved in the indole pathway of the gut microbial Trp metabolism [12]. In contrast, another Trp metabolite, Kyn, was increased in the feces of Abxtreated mice (Fig. 4(c)). Xanthurenic acid was also increased in Abx-treated mice, which was probably excreted by the host, because the corresponding enzymes were found in the host rather than in the microbiota [12]. Moreover, Abx treatment by oral gavage for four weeks did not affect the body weight of mice (Fig. S3(c)), indicating that the amount of Trp intake from the daily diet was almost equal for the Abx-treated and the untreated mice. These data revealed that Abx facilitated Trp excretion through feces and changed the gut microbial Trp metabolism, with decreased production of several indole derivatives and increased production of Kyn.
Thus, Abx reshaped the gut microbial metabolism with increased Kyn production and reduced Trp excretion, which might allow more Kyn and Trp to be accessible to the host and regulate the host’s metabolic immune responses.
《Fig. 4》
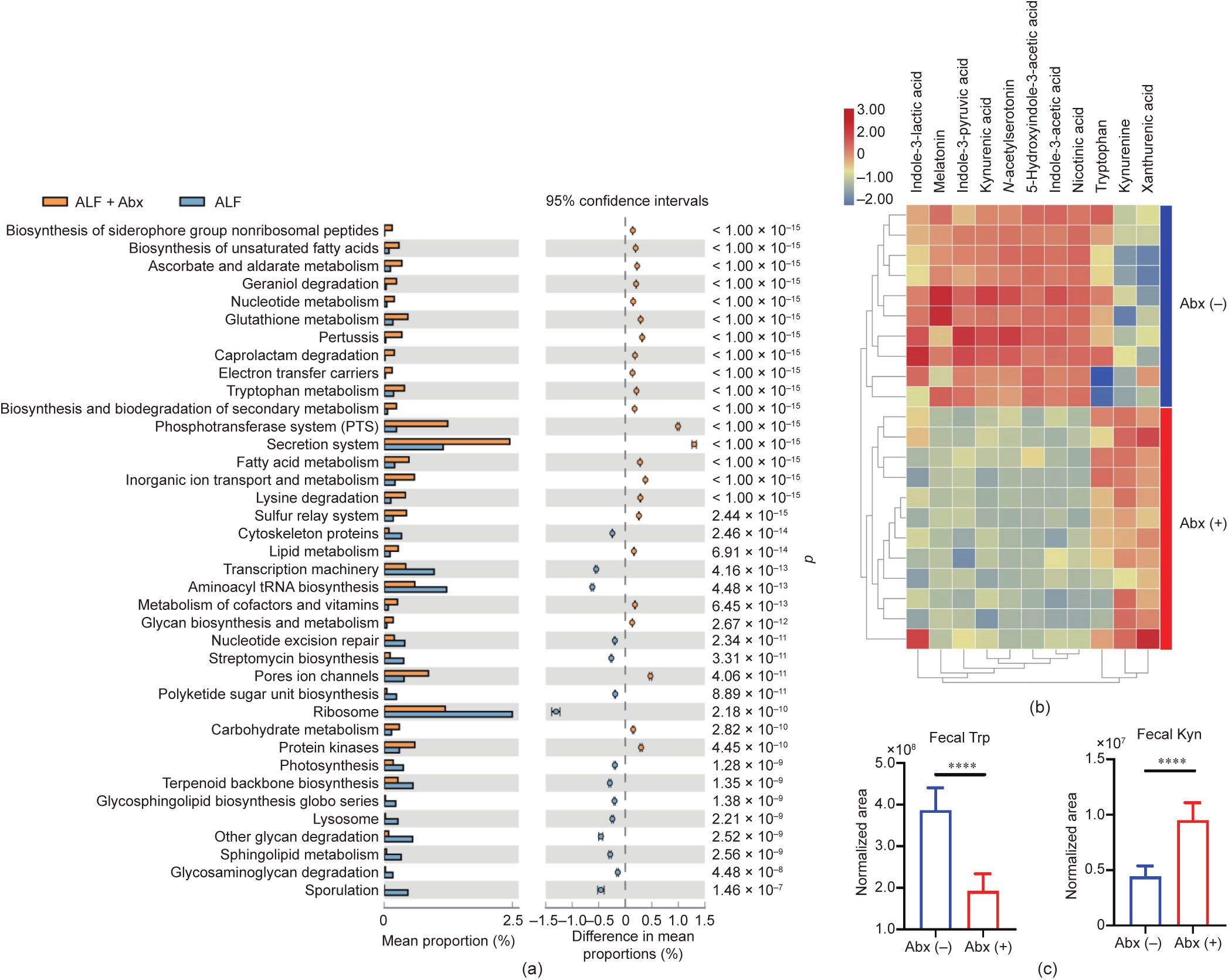
Fig. 4. Antibiotics reshaped the gut microbial metabolism with increased Kyn production and reduced Trp excretion. (a) Predicted metagenome functional content at KEGG level 3 (n = 9–11 per group). (b) Heatmap of fecal Trp and its metabolites identified by global metabolic profiling (n = 10–12 per group). (c) Alteration of Trp and Kyn in the feces between Abx-treated (+) and untreated (–) mice (n = 10–12 per group). Data is shown by: (a) mean proportion and its 95% confidence intervals only with the difference of mean proportions > 0.12%, a ratio of proportion > 2, and p < 0.01 between two groups using a Welch’s t test; and (c) mean ± SD and ****p < 0.0001 using an unpaired t test.
《3.5. Serum Kyn and Kyn/Trp ratio were increased in Abx-treated ALF mice》
3.5. Serum Kyn and Kyn/Trp ratio were increased in Abx-treated ALF mice
The major pathway of the hepatic Trp metabolism is the Kyn pathway, which is initiated by two enzymes: tryptophan 2,3- dioxygenase (TDO) and indoleamine 2,3-dioxygenase 2 (IDO2) [25]. Hepatic gene expressions of Tdo and Ido2 were downregulated in ALF mice compared with Con mice and were upregulated in ALF + Abx mice compared with ALF mice (Fig. 5(a)), suggesting that the ability to metabolize Trp was reduced in the liver of ALF mice but improved in that of ALF + Abx mice. UPLC–MS was used to detect Trp and Kyn levels in the liver tissue and serum. Although there was no statistical difference, the Trp levels of liver and serum tended to be higher in ALF mice than in Con mice (Fig. 5(b)), probably due to the elevation of plasma-free Trp. It has been found that plasma-free Trp is increased in fulminant liver failure because of low circulating albumin and high non-esterified fatty acid (NEFA) levels [26–28]. Liver Kyn was found to be significantly increased in ALF mice but not in ALF + Abx mice compared with Con mice (Fig. 5(c)), which might be a compensative response to counteract the adverse effect of local severe inflammation in the liver of ALF mice. Serum Kyn was significantly increased in ALF + Abx mice compared with Con mice and ALF mice (Fig. 5(c)), which was positively associated with the increased fecal Kyn in Abx-treated mice (Fig. 5(d)). The ratio of plasma Kyn/Trp reflects systemic IDO activity, and IDO-mediated Trp degradation is a potent immunosuppressive mechanism [25]. In this study, the serum Kyn/Trp ratio was increased in ALF + Abx mice compared with Con mice and ALF mice (Fig. 5(e)), suggesting that systemic IDO activity was increased in ALF + Abx mice.
The Abx-induced compositional changes in the gut microbiota shifted the Trp metabolism of the host, leading to an increased serum Kyn and Kyn/Trp ratio in ALF + Abx mice, which may inhibit the inflammatory response and protect mice from ALF.
《Fig. 5》
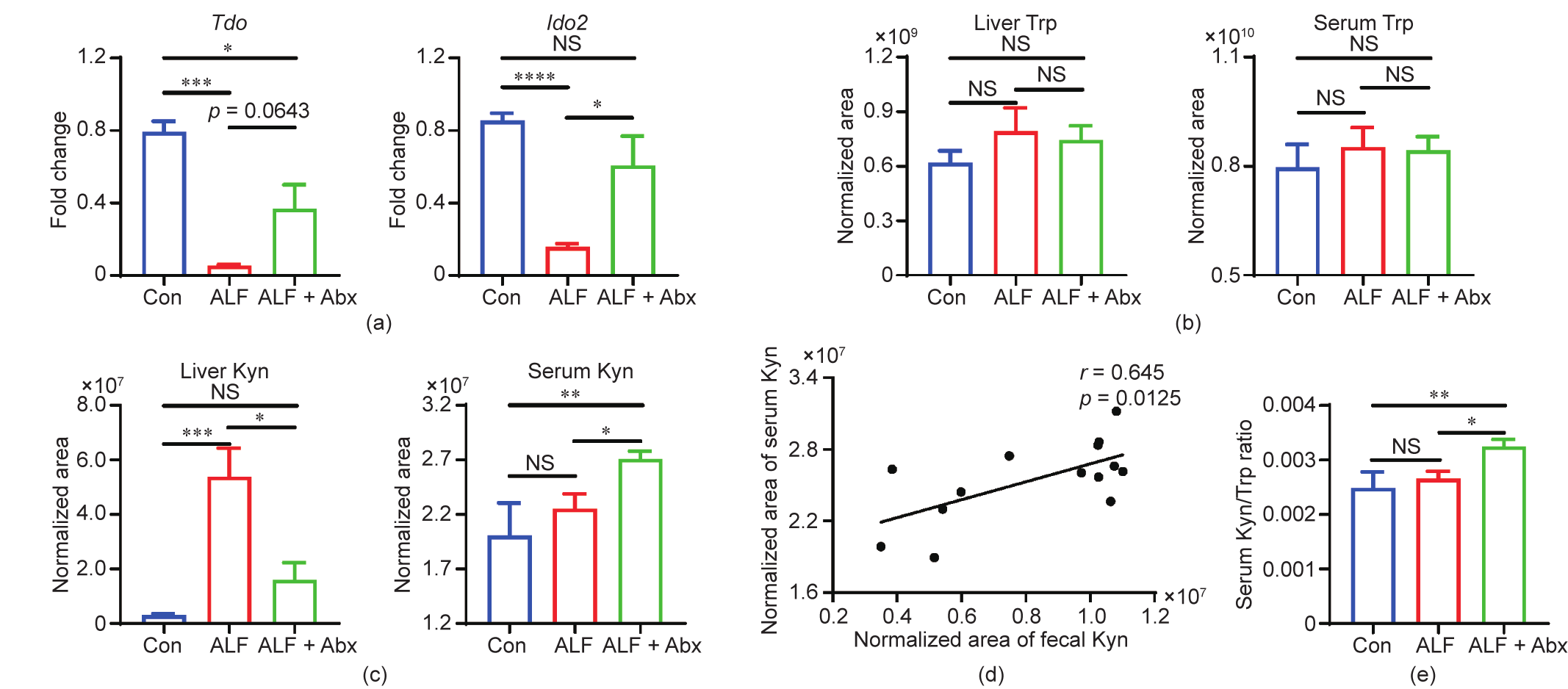
Fig. 5. Serum Kyn and Kyn/Trp ratio were increased in Abx-treated ALF mice. (a) Hepatic gene expression of Tdo and Ido2 (n = 5–6 per group). (b) Alteration of Trp in the liver and serum (n = 5–9 per group). (c) Alteration of Kyn in the liver and serum (n = 5–9 per group). (d) Linear correlation analysis between fecal Kyn and serum Kyn (n = 14). (e) Ratio of Kyn/Trp in the serum (n = 5–9 per group). Data is shown by mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using ANOVA, Kruskal–Wallis, or Pearson test.
《3.6. The AhR signal was partly reverted in Abx-treated ALF mice》
3.6. The AhR signal was partly reverted in Abx-treated ALF mice
The anti-inflammatory and immunosuppressive effects of Kyn are largely mediated by its function as a ligand of AhR, which is a transcription factor that regulates local and systemic immune responses [12,14,29]. The hepatic gene expression of Ahr was downregulated in ALF mice, but partly reverted in ALF + Abx mice, compared with Con mice (Fig. 6(a)), suggesting that AhR is involved in the pathophysiology of ALF. Inactive AhR is localized in the cytoplasm; once ligand binding triggers AhR and exposes its nuclear localization site, the AhR complex transfers into the nucleus and interacts with the AhR nuclear translocator (ARNT), resulting in the transcription of multiple target genes, such as xenobiotic metabolic enzyme cytochrome P450 family-1 subfamily-A polypeptide-1 (CYP1A1) [30]. The hepatic gene expression of Arnt and the AhR target gene Cyp1a1 were also upregulated in ALF + Abx mice in comparison with ALF mice (Fig. 6(a)), suggesting that the AhR signal was partly reverted in ALF + Abx mice. In order to determine whether the observed upregulated gene expression of Ahr resulted in AhR activation, immunofluorescence staining of AhR was performed. The immunofluorescence intensity of hepatic AhR was significantly reduced in ALF mice in comparison with Con mice, and was increased in ALF + Abx mice in comparison with ALF mice (Fig. 6(b) and Fig. S4(a) in Appendix A). AhR activation can also promote cell proliferation by modulating growth factors and the cell cycle [31]. Therefore, the expression level of hepatic PCNA was assessed by means of immunohistochemical staining. There was an obvious difference in proliferation state between ALF mice and ALF + Abx mice (Fig. 6(c)). The proliferation index was decreased in ALF mice but significantly increased in ALF + Abx mice in comparison with Con mice (Fig. S4(b)); this suggests that cell proliferation was stimulated in ALF + Abx mice, which may be due to Kyn dependent AhR activation. These data suggest that the AhR signal was partly reverted in ALF + Abx mice, and the consequent immunosuppression and repair process could be the underlying mechanism causing liver injury to be attenuated in Abx-treated ALF mice.
《Fig. 6》

Fig. 6. The AhR signal was partly reverted in Abx-treated ALF mice. (a) Hepatic gene expression of Ahr, Arnt, and Cyp1a1 (n = 10–12 per group). (b) Representative images of hepatic AhR immunofluorescence staining (green). (c) Representative images of hepatic PCNA immunohistochemistry. Data is shown by mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using Kruskal–Wallis test.
《3.7. The TLR4 signal was impaired in Abx-treated ALF mice》
3.7. The TLR4 signal was impaired in Abx-treated ALF mice
For the ALF model we used, pathophysiology starts with LPS binding to TLR4, and D-Gal sensitizes the rodents to LPS [32]. LPS recognition at the cell surface is a biological process that not only involves TLR4, but also requires LPS binding protein (LBP) and CD14. LBP is able to mediate LPS internalization, while cluster of differentiation 14 (CD14) is primarily responsible for LPS/TLR4 activation signal transduction [33]. Although the hepatic gene expression of Tlr4 and Cd14 showed no difference between ALF mice and ALF + Abx mice, the hepatic gene expression of Lbp was upregulated in ALF + Abx mice compared with ALF mice (Fig. 7(a)), which probably mediated LPS internalization to decrease inflammatory responses. Moreover, hepatic TLR4 immunoreactivity was decreased in ALF + Abx mice in comparison with ALF mice (Fig. 7(b) and Fig. S5(a) in Appendix A). TLR4 signaling is important for the recruitment and activation of macrophages. Immunofluorescence staining of hepatic F4/80, a major macrophage marker, indicated that the recruitment and activation of macrophages were inhibited in ALF + Abx mice compared with ALF mice (Fig. 7(c)). This was confirmed by a decreased integrated fluorescence density of F4/80 in ALF + Abx mice compared with ALF mice (Fig. S5(b)). The hepatic expression of 24-dehydrocholesterol reductase gene (Dhcr24), which inhibits the production of TNF-α by macrophages, was upregulated in ALF + Abx mice compared with ALF mice, while the hepatic gene expression of both Tnf-α and its downstream gene, NOD-like receptor family pyrin domain containing 3 gene (Nlrp3), which codes for pro-inflammatory mediators, was down-regulated in ALF + Abx mice compared with ALF mice (Fig. 7(d)). Therefore, the TLR4 signal was impaired in ALF + Abx mice, which may be the underlying mechanism of immunosuppression due to Kyn-induced AhR activation.
《Fig. 7》

Fig. 7. The TLR4 signal was impaired in ALF mice with antibiotics pretreatment. (a) Hepatic gene expression of Lbp, Tlr4, and Cd14 (n = 10–12 per group). (b) Representative images of hepatic TLR4 immunofluorescence staining (green). (c) Representative images of hepatic F4/80 immunofluorescence staining (red). (d) Hepatic gene expression of Dhcr24, Tnf-α, and Nlrp3 (n = 10–12 per group). Data is shown by mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using ANOVA or Kruskal–Wallis test.
《3.8. The AhR agonist partly mimicked while the antagonist incompletely counteracted the protective effects of antibiotics on ALF mice》
3.8. The AhR agonist partly mimicked while the antagonist incompletely counteracted the protective effects of antibiotics on ALF mice
AhR-targeted treatment strategies were used to study the role of AhR in ALF. The serum AST was increased and the serum ALT tended to increase in ALF + Abx + AhR antagonist mice compared with ALF + Abx mice (Fig. 8(a)), suggesting that the AhR antagonist aggravated liver damage in ALF + Abx mice. Furthermore, the AhR antagonist accelerated the death of Abx-treated mice at the early stage after ALF induction, but could not completely counteract the effect of Abx pretreatment on the survival of ALF mice in terms of the final outcome (Fig. 8(b)). Hence, the direct effects of the AhR agonist on ALF were evaluated. The serum ALT was decreased but the serum AST was not decreased in ALF + AhR agonist mice compared with ALF mice (Fig. 8(c)). In addition, the AhR agonist prolonged the survival time of ALF mice, but did not improve their survival rate (Fig. 8(d)). The AhR-targeted gene Cyp1a1 was upregulated, the gene expression of Lbp showed an increasing trend, and the gene expression of Tnf-α was downregulated in ALF + AhR agonist mice compared with ALF mice (Fig. 8(e)). Taken together, these findings indicate that the AhR agonist could partly mimic and the AhR antagonist incompletely counteracted the protective effects of Abx pretreatment on ALF mice, suggesting that the effects of the gut microbiota post-Abx treatment on ALF were mediated, at least in part, by the AhR pathway.
《Fig. 8》

Fig. 8. The AhR agonist partly mimicked while the antagonist incompletely counteracted the protective effects of antibiotics on ALF mice. (a) Serum ALT and AST levels (n = 9–10 per group). (b) Cumulative survival analysis was determinead with a Kaplan–Meier curve (n = 9–10 per group). (c) Serum ALT and AST levels (n = 8–10 per group). (d) Cumulative survival analysis was determined with a Kaplan–Meier curve (n = 8–10 per group). (e) Hepatic gene expression of Cyp1a1, Lbp, and Tnf-α (n = 4–5 per group). Data is shown by mean ± SEM. *p < 0.05, **p < 0.01, ***p < 0.001, and ****p < 0.0001 using ANOVA, Kruskal–Wallis, or log-rank test.
《4. Discussion》
4. Discussion
As a significant player in liver physiology and pathophysiology, the gut microbiota has become an attractive research field [34]. Due to its specific position, the liver is constantly exposed to gutderived endotoxins, antigens, and microbial metabolites [35]. A diverse set of gut-derived metabolites, which can shape the host immune system, allow the gut microbiota to exert multiple effects at a distance. SCFAs, polyamines, and Trp metabolites are three of the currently most-studied categories of metabolites produced by the gut microbiota [5]. Compositional changes in the gut microbiota will lead to alteration of the gut microbial metabolism, which may impact the host’s immune responses. Under normal conditions, immunological tolerance maintains hepatic homeostasis. However, when equilibrium is broken and the immune system is excessively activated by internal and external factors, acute liver injury will happen. Although previous studies have demonstrated that gut-derived endotoxins and antigens play a universal role in liver injury [2], the effects of the gut microbial metabolism on liver injury remain unclear. In this study, we reported a previously unappreciated role of the gut microbiota-controlled Trp metabolism in the regulation of immune responses in ALF. Abx enhanced the Kyn production of the gut microbial Trp metabolism, allowed more Trp to be accessible to the host, and increased serum Kyn, which improved D-Gal/LPS-induced ALF in C57BL/6 mice, probably through the AhR signal.
Abx pretreatment improved the survival of D-Gal/LPS-induced ALF in C57BL/6 mice. Abx pretreatment by gavage for four weeks decreased gut bacterial load, which limited gut-derived LPS. Gutderived LPS has been identified as a critical cofactor in the liver injury model established by carbon tetrachloride (CCl4), alcohol, D-Gal, or acetaminophen [2]. However, we used D-Gal combined with LPS to induce ALF, which could partly counteract the effect of the Abx-mediated decrease of gut-derived LPS; furthermore, Abx did not cause a complete clearance of bacteria due to the emergence of antibiotic-resistant bacteria. Therefore, the role of gut microbiota post-Abx treatment in ALF requires further study.
Abx treatment for four weeks expanded specific indigenous taxa. At the genus level, Enterobacteriaceae unclassified, Escherichia–Shigella, and Morganella accounted for the majority of gut microbiota. Moreover, all of them are responsible for tryptophanase A (TnaA) encoding and indole production [36]. Indole is one of the microbial metabolites produced from Trp. Trp is one of nine essential aromatic acids that are critical for protein synthesis, which must rely on exogenous intake; it also serves as the substrate for the generation of several bioactive compounds. The intestinal tract plays an important role in Trp metabolism, which follows three major metabolic fates: the serotonin pathway, Kyn pathway, and indoles pathway [12]. In general, the gut microbiota mainly metabolizes Trp into indole and indole derivatives [37]. However, the identified indole derivatives were decreased in the feces of Abx-treated mice, whereas the mean proportion of the gut microbial Trp metabolism was increased and Kyn was increased. These findings indicate that Abx shunted the gut microbial Trp metabolism toward Kyn production. Interestingly, Trp was also decreased in the feces of Abx-treated mice, suggesting that Abx reduced Trp absorption by the gut microbiota. Previous studies have found that both Abx-treated mice and germ-free mice have a high circulating level of Trp, which can be normalized by FMT [10,11]. Therefore, the gut microbiota could not only metabolize Trp, but also regulate Trp availability to the host.
Under normal conditions, after the intake of Trp, a portion is utilized by the gut microbiota and the rest is absorbed by the host. Trp has low tissue storage and is predominantly bound to albumin in the circulation; free Trp that can be degraded only accounts for 5%–10% of the total Trp [13,25]. The major metabolic pathway of Trp is the Kyn pathway. The liver contains a set of enzymes required for any branch of the Kyn pathway, which has a central role in the regulation of the systemic level of Trp [13]. TDO and IDO2 mediate the first step of Trp metabolism in the liver, and hepatic TDO can be directly induced by its substrate Trp [25]. In this study, the hepatic expression of Tdo was increased in ALF + Abx mice in comparison with ALF mice, indicating that Trp availability and metabolism were increased in ALF + Abx mice. In addition, the hepatic expression of Ido2 was upregulated in ALF + Abx mice compared with ALF mice. The serum Kyn/Trp ratio, which reflects systemic IDO activity, was also increased in ALF + Abx mice compared with Con mice and ALF mice. IDO is highly expressed in immune cells, and Trp metabolism via IDO is an important step in metabolic immune regulation [13]. One of the proposed underlying mechanisms is the production of Trp metabolites, which have immune regulation activity, such as Kyn [38]. Previous studies have showed that Kyn inhibits the function of natural killer (NK) cells and antigen presenting cells (APCs), such as monocytes, macrophages, and dendritic cells [39,40]. Furthermore, Kyn suppresses T cell proliferation leading to the death of T cells, and Kyn production in dendritic cells contributes to the proliferation of regulatory T cells [41,42]. Kyn also induces endotoxin tolerance in dendritic cells [43]. The immunosuppressive effects of Kyn in the regulation of inflammation are, at least in part, mediated by its function as a ligand of AhR, an important transcription factor that is widely expressed in immune cells and that controls local and systemic immune responses [29].
AhR is involved in the xenobiotic metabolism of foreign substances, which controls the transcription of various target genes such as cytochrome P450-dependent monooxygenases CYP1A1 and cytochrome P450 family-1 subfamily-A polypeptide-2 (CYP1A2) [30]. In addition to mediating the toxicity of environmental pollutants, AhR has immunological importance [44]. AhR plays an important role in the regulation of immune responses in health and diseases, such as infection, nervous system diseases, inflammatory bowel disease, autoimmune disease, and metabolic syndrome [45–47]. AhR interacts with other transcription factors, such as nuclear factor (NF)-κB and c-Maf, to modulate the transcription of their targeted genes [48]. In addition to genomic mechanisms, AhR regulates immune responses by non-genomic signaling. AhR-controlled c-SRC activation plays a key role in the control of a disease tolerance defense pathway and in limiting macrophage responses to inflammatory stimuli [49,50]. However, the role of AhR signaling in ALF is still unclear. We observed that Ahr gene expression was significantly downregulated in mice with D-Gal/LPS-induced ALF, which was attenuated in ALF + Abx mice, suggesting that the effect of Abx was at least partly AhR dependent. The upregulated Arnt and Cyp1a1 gene expression in ALF + Abx mice compared with ALF mice indicated the activation of AhR in ALF + Abx mice, which was confirmed by increased AhR immunofluorescence activity. The immune shift mediated by AhR activation could prevent uncontrolled inflammatory responses. In addition, AhR has dual functions in the regulation of cell proliferation and survival, which may be due to differences in dosage and time frame of the ligand, cell types, or whether the experiment is performed in vivo or in vitro [31]. Nevertheless, AhR is important for tissue regeneration by modulating receptor expression, participating in growth factor signaling, promoting anti-apoptosis, regulating the cell cycle, and promoting cytokine expression [31]. In this study, a significant difference was observed in liver cell proliferation, as measured by PCNA immunohistochemistry, between Abx-treated and untreated mice with ALF, indicating that the hepatic repair process was facilitated by Abx-induced increased Kyn production by the microbiota and host. Moreover, as an AhR agonist, indole potentially leads to the impairment of LPS signaling by reducing the mRNA level of Cd14 [51]. LPS recognition at the cell surface is a biological process involving three key proteins: TLR4, CD14, and LBP. LBP is able to mediate LPS internalization, while CD14 is primarily responsible for TLR4 activation signal transduction [33]. At a low level, LBP enhances the LPS-induced inflammatory response, whereas a higher level of LBP neutralizes LPS and alleviates the LPS-induced activation of TLR4 [52]. In this study, the hepatic gene expression of Lbp was upregulated in ALF + Abx mice, which probably mediated LPS internalization or neutralization and then impaired the TLR4 signal. Therefore, both AhRmediated immune suppression and cell proliferation may be involved in the protective effects of the Abx-induced activation of the Kyn pathway. Moreover, the AhR antagonist incompletely counteracted the protective effects of Abx pretreatment on ALF, which could be due to the size of the experiment, the time frame and dosage of the ligand, the method of administration, and offtarget effects. However, the AhR agonist improved liver function and prolonged the survival time of mice with ALF. Therefore, in addition to a broad decrease in bacterial load with limited LPS, gut microbiota-controlled Trp metabolism decreased the host susceptibility to liver injury by modulating the AhR signal.
While ALF is an important clinical problem, the necessity for pretreatment with Abx largely diminishes its therapeutic potential. Further interventional studies should evaluate the treatment effects of direct Kyn supplementation or AhR-targeted therapeutic strategies after the onset of ALF. However, in the case of liver transplantation, Abx recipient pretreatment alleviates hepatic ischemia reperfusion injury and improves the outcome of orthotropic liver transplantation in mice and humans [16]. Although it is still unclear whether Abx-mediated Kyn–AhR activation is involved in the underlying mechanisms, Abx treatment may be appropriate for advanced ALF patients who must receive a liver transplant but are waiting for a liver donor. However, it should be noted that antibiotic-resistant bacteria and economic efficiency discourage the overuse of Abx; therefore, further studies are needed in appropriate Abx regimens, such as rifaximin, and complementary gut microbiota modification strategies, such as probiotics, prebiotics, and postbiotics. Furthermore, AhR has been implicated in chronic diseases such as non-alcoholic fatty liver disease (NAFLD), alcoholic liver disease (ALD), and liver fibrosis [46,47,53]. As these liver injuries last over a longer time frame than ALF, it is possible that gut microbiota modification can be used to therapeutically boost AhR activity and alleviate liver injury after diagnosis.
This study has a few limitations. Although immunohistochemical and immunofluorescence staining, combined with real-time quantitative PCR, are useful to determine protein expression levels, the results of Western blot tests are likely to be more persuasive. In addition, only an untargeted approach was used to detect metabolites, although a targeted approach is preferable for confirmation. Furthermore, the use of a gene knockout animal is a more ideal method for studying the function of AhR in ALF. These lessons should be noted and overcome in future research.
In conclusion, this study increases the current understanding of how Abx-mediated gut microbiota can affect susceptibility to liver diseases by regulating the host metabolic immune response. Abx shunted the gut microbial Trp metabolism toward Kyn production and liberated more Trp to the host; this increased the immunosuppressive metabolite Kyn, which improved D-Gal/LPS-induced ALF, probably through the AhR signal. This study demonstrates that the gut microbiota-controlled Trp metabolism regulated susceptibility to liver diseases by engaging the host receptor AhR. It also emphasizes that further studies on gut microbiota–host Trp cometabolism will provide promising therapeutic strategies for liver diseases.
《Acknowledgments》
Acknowledgments
This work was supported by the Major Program of the National Natural Science Foundation of China (81790630 and 81790633), the Sino-German Center for Research Promotion (GZ1546), and the Chinese Academy of Medical Sciences (CAMS) Innovation Fund for Medical Sciences (2019-I2M-5-045).
《Compliance with ethics guidelines》
Compliance with ethics guidelines
Zhipeng Zheng, Li Wu, Yuqiu Han, Jun Chen, Shuai Zhu, Yuanyuan Yao, Baohong Wang, and Lanjuan Li declare that they have no conflict of interest or financial conflicts to disclose.
《Appendix A. Supplementary data》
Appendix A. Supplementary data
Supplementary data to this article can be found online at https://doi.org/10.1016/j.eng.2020.12.013.













 京公网安备 11010502051620号
京公网安备 11010502051620号




